Abstract
The budding yeast Saccharomyces cerevisiae is unable to incorporate exogenous nucleosides into DNA. We have made a number of improvements to existing strategies to reconstitute an efficient thymidine salvage pathway in yeast. We have constructed strains that express both a nucleoside kinase as well as an equilibrative nucleoside transporter. By also deleting the gene encoding thymidylate synthase (CDC21) we have constructed strains that are entirely dependent upon exogenous thymidine for viability and that can grow with normal kinetics at low thymidine concentrations. Using this novel approach, we show that depletion of a single deoxyribonucleoside causes reversible arrest of cells in S phase with concomitant phosphorylation and activation of the S phase checkpoint kinase, Rad53. We show that this strain also efficiently incorporates the thymidine analogue, BrdU, into DNA and can be used for pulse–chase labelling.
INTRODUCTION
The budding yeast, Saccharomyces cerevisiae, has been extremely useful for studying DNA replication (1). Replication initiates from short, well-defined replication origins and is amenable to sophisticated genetic manipulation. For many DNA replication experiments, it would also be useful to be able to label yeast DNA using precursor nucleosides. Unfortunately, because budding yeast is unable to ‘salvage’ nucleosides from the environment, a system for efficient labelling of yeast DNA with precursors has not been available. There are two primary reasons for the inability of yeast to salvage nucleosides. First, yeast lack the enzyme required to phosphorylate deoxyribonucleosides, converting them to 5′ deoxyribonucleoside monophosphates. Secondly, they lack the capacity for mediated uptake of exogenous deoxyribonucleosides.
The inability to phosphorylate deoxyribonucleosides has previously been addressed by expressing the thymidine kinase (TK) from Herpes Simplex Virus (HSV). When HSV-TK is expressed, yeast can grow even if de novo dTTP synthesis is inhibited either with drugs or with a temperature sensitive mutant, if a high level of thymidine (2 mM) is supplied in the medium (2,3). Introduction of multiple copies of HSV-TK allows incorporation of detectable levels of the thymidine analogue, 5-bromodeoxyuridine (BrdU) into DNA (3,4).
The need for high thymidine concentrations, however, makes this approach impractical for incorporation of radiolabelled precursors. Moreover, because there is no active import of nucleosides from the medium, uptake is slow, making ‘pulse–chase’ experiments impractical. Mutants that allow thymidine uptake have been isolated (5,6); however, such tup or tut mutants (e.g. tup1) generally have pleiotropic phenotypes (6,7).
In mammalian cells, a series of transporters exist which mediate uptake of deoxyribonucleosides. These include equilibrative transporters, which facilitate diffusion down a concentration gradient, and concentrative transporters, which work against a concentration gradient (8). Though both might be useful to reconstitute a salvage pathway, we have chosen to use the equilibrative transporter because it should allow greater flexibility in altering intracellular dNTP pools by altering extracellular nucleoside concentrations. Previous work has shown that yeast strains that express both HSV-TK and the human equilibrative nucleoside transporter (hENT) can grow even when de novo dTTP synthesis is inhibited with drugs (9). In this study we have extended this work to construct an efficient salvage pathway for dTTP biosynthesis in yeast. This was initially accomplished by expressing hENT1 (9,10) with HSV-TK. During the course of this work we found that the Drosophila melanogaster deoxyribonucleoside kinase (DmdNK) (11,12), which is faster and has a broader specificity than HSV-TK, could also be used. By deleting the CDC21 gene, which encodes thymidylate synthase (13,14) and is essential for de novo dTTP biosynthesis, we have constructed strains that rely entirely on exogenous thymidine for growth. We show that thymidine depletion provokes an S phase arrest accompanied by activation of the S phase checkpoint. This arrest is rapidly reversed by addition of thymidine. These results provide a direct link between nucleotide depletion and checkpoint activation. We have also found that these strains are able to take up BrdU and incorporate it into DNA as the sole ‘thymidine’ source without accompanying Rad53 activation, providing a means of labelling DNA in vivo in yeast. We show that this procedure can be used for non-radioactive pulse–chase labelling of nascent DNA. Our results also represent a platform for screening novel pairs of deoxyribonucleoside kinase mutants and analogues/drugs with a potential for gene therapy.
MATERIALS AND METHODS
Strains and media
Strains W303-1a (MATa ade2-1 ura3-1 his3-11,15 trp1-1 leu2-3,112) and W303 (MATa/MATα ade2-1/ade2-1 ura3-1/ura3-1 his3-11,15/his3-11,15 trp1-1/trp1-1 leu2-3,112/leu2-3,112) were used in this study. Yeast strains were grown in rich medium YP (1% yeast extract, Difco, plus 2% bacto peptone, Difco), or in minimum medium YNB (0.67% yeast nitrogen base, Difco, adenine 40 µg/ml, histidine 40 µg/ml, leucine 80 µg/ml, tryptophan 40 µg/ml and uracil 40 µg/ml) supplemented with glucose or galactose to a final concentration of 2% and with 5% casamino acids, Difco. Agar was added for growth on plates to 2% final concentration. BrdU was obtained from Sigma. Methotrexate and sulfanilamide were added to final 50 µg/ml and 6 mg/ml, respectively. Diploid strains were sporulated on Rich Sporulation Medium containing galactose 0.1% and G418 200 µg/ml if required for Kanamycin resistance selection. All experiments with yeast were carried out at 30°C. GAL1-10 promoter was amplified by PCR and subcloned into plasmids pRS304, pRS305, pRS424 and pRS425 using EcoRI and BamHI sites. HSV-TK gene was amplified by PCR using the following oligos: GCTAggatccGCCATCATGGCCTCGTACC (BamHI site and ATG are both in bold) and GCTAgcggccgcATCTAGTCAGTTAGCCTCC (NotI site in bold) on plasmid pGT60lacZ (InvivoGen). The PCR product was digested with BamHI + NotI and subcloned into pRS304-GAL1-10 and pRS424-GAL1-10 vectors.
A BamHI/NotI fragment containing the DmdNK gene was obtained from the yeast expression vector pYES2 (Invitrogen) expressing the DmdNK gene and subcloned into pRS305-GAL1-10 (12).
hENT1 gene was amplified by PCR using the following oligos: GCTggatccATGACAACCAGTCACCAGCCT (BamHI site in bold is immediately followed by the ATG of the gene) and GCTgcggccgcTCACACAATTGCCCGGAA (NotI site in bold) from plasmid pYhENT1 (9). The PCR product was digested with BamHI + NotI and subcloned into pRS305-GAL1-10 and pRS425-GAL1-10 vectors. To delete the CDC21 gene, a fragment from the plasmid pUG6 containing the kanMX cassette was amplified by PCR, using the following oligonucleotides: atacgtaagtacatgattttgtttctcctcgtgctgtcaaCAGCTGAAGCTTCGTACGC (5′ end of the KAN gene in upper case and 3′ end reverse of the CDC21 gene, just after the stop in bold) and aaagtatcaaggagagagcttcataacagaacggtacaggCATAGGCCACTAGTGGATCTG (3′ end of the KAN gene in uppercase and 5′ end direct of the CDC21 gene just in front of the ATG). GFP plasmid pAFS91 was previously described (15) and integrated in yeast at the HIS3 locus following a StuI digest. GFP-Tubulin was then detected in live cells. One millimetre of cells was harvested from a YPGal culture with a density of 5–10 × 107 cells/ml. Cells were washed with 1 ml water and resuspended in 5 µl water, and 1 µl of cells was then deposited on a glass slide. One microlitre of mounting solution (90% glycerol, 0.5 µg/ml DAPI) was then added directly on the slide and mixed rapidly. Two microlitres of a 2% low melting agarose (GTG Seakem) solution in water maintained at 42°C was added on the top. The mix was then covered with a cover slip before direct observation under the microscope.
Blotting for BrdU-labelled DNA
DNA from frozen cell pellets was purified using phenol extraction, quantified on gel and run on a 1.2% alkaline agarose (16,17). DNA was transferred from gels to Hybond N+ membranes according to the manufacturer’s instructions. The membrane was blocked with 5% dry milk in Tris-buffered saline containing 0.1% Tween 20. BrdU-labelled DNA was detected using anti-BrdU antibody (Becton-Dickinson, 1/1000) and an anti-mouse antibody coupled to horseradish peroxidase. The product of the reaction was detected with enhanced chemiluminescence (Amersham) according to the manufacturer’s instructions.
Other methods
BrdU (Sigma) detection in yeast cells was performed as described previously (18), using a mouse monoclonal anti-BrdU antibody (Becton-Dickinson) and an Alexa Fluor 488 goat anti-mouse (Molecular Probes). Samples for flow cytometric analysis were collected and processed as described (19).
RESULTS AND DISCUSSION
Growth of strains expressing HSV-TK and hENT1
During de novo dTTP synthesis, dTMP is generated by methylation of dUMP. This reaction is catalysed by thymidylate synthase, encoded by the CDC21 gene and requires N5,N10 methylenetetrahydrofolate which donates the methyl group and is concomitantly converted to dihydrofolate (14). The regeneration of N5,N10 methylenetetrahydrofolate from dihydrofolate involves reduction of dihydrofolate to tetrahydrofolate by dihydrofolate reductase (DHFR) and generation of N5,N10 methylenetetrahydrofolate from tetrahydrofolate by serine transhydroxymethylase. Several widely used drugs block dTMP synthesis by interfering with the synthesis of N5,N10 methylenetetrahydrofolate: mimosine inhibits serine transhydroxymethylase (20), the folate analogue methotrexate (amethopterin) inhibits DHFR (21), while sulphanilamide competes with p-aminobenzoic acid to inhibit dihydropteroate synthetase (22), thus blocking the synthesis of 7,8-dihydropterate, a precursor of dihydrofolate.
To reconstitute a salvage pathway in yeast, we expressed both HSV-TK and hENT1 from the GAL1-10 promoter. Each gene was either integrated in the chromosome as a single copy, or expressed from a multicopy plasmid. The different strains were assessed for growth on minimum galactose medium plates in the presence of methotrexate and sulfanilamide, together with various amounts of thymidine. Results are shown in Table 1. Strains expressing at least one copy of each gene were able to grow in the presence of the drugs in a thymidine-dependent manner. A strain expressing HSV-TK from a multicopy plasmid was able to grow without hENT1; however, it required a very high thymidine concentration (2 mM). In strains expressing hENT1, there was no difference in growth between HSV-TK expression from a multicopy plasmid compared with HSV-TK expression as a single chromosomal copy, whereas hENT1 overexpression from a multicopy plasmid allowed the yeast to grow with much lower amounts of thymidine (dT) in the medium (from 1 µM dT in the medium compared with 100 µM dT). Because the growth of yeast requires exogenous thymidine when the de novo pathway is inhibited by drugs, these results show that a thymidine salvage pathway is functioning consistent with previous results (9). The efficiency of the salvage pathway is improved with increased expression of hENT1, presumably because thymidine uptake is modulated accordingly. Because the copy number of multicopy ARS plasmids varies in the yeast population, however, the extent of gene overexpression will be heterogeneous within the cells of a population. For this reason, the strain with single integrated copies of each gene was selected for further study.
Table 1. Growth of hENT1- and TK-expressing strains.
| Strain | dT concentration | |||||
|---|---|---|---|---|---|---|
| 2 mM | 100 µM | 10 µM | 5 µM | 1 µM | 0 | |
| W3031a | – | – | – | – | – | – |
| W3031a | ||||||
| GAL-TK multicopy | + | – | – | – | – | – |
| W3031a | ||||||
| GAL-hENT1 multicopy | – | – | – | – | – | – |
| W3031a | ||||||
| GAL-TK multicopy | ||||||
| GAL-hENT1 multicopy | + | + | + | + | + | – |
| W3031a | ||||||
| GAL-TK multicopy | ||||||
| GAL-hENT1 integrated | + | + | – | – | – | – |
| W3031a | ||||||
| GAL-TK integrated | ||||||
| GAL-hENT1 integrated | + | + | – | – | – | – |
CDC21 deletion is rescued on thymidine-containing medium
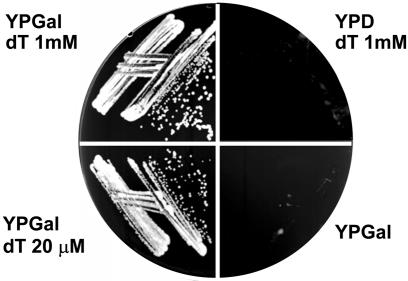
Methotrexate and sulfanilamide can have pleiotropic effects on cell metabolism by interfering with folate metabolism. Therefore, we eliminated the de novo pathway for dTTP biosynthesis genetically by deleting CDC21, the gene encoding thymidylate synthase. To accomplish this, one copy of CDC21 was deleted using the kanamycin resistance cassette in the diploid strain W303. Both the GAL-TK and GAL-hENT1 constructs were then integrated into this diploid strain. After sporulation, spores harbouring the cdc21 deletion were selected on YPGal medium containing both kanamycin and thymidine (100 µM). All the germinating spores were LEU+, TRP+, indicating that they also carried both the GAL-TK and GAL-hENT1 constructs. Since dTTP biosynthesis can only occur via the salvage pathway, growth of the resulting strain (YLVA10) is strictly dependent on the presence of thymidine and galactose in the medium. Figure 1 shows that cell growth required both galactose and thymidine in the medium and cells were able to grow well with a thymidine concentration as low as 20 µM in the medium (Fig. 1).
Figure 1.
Reconstitution of an efficient salvage pathway for dTTP biosynthesis in yeast. Cells expressing HSV-TK and hENT1, and deleted for the CDC21 gene, were streaked on YPGal (inducing conditions) or YPD (repressing conditions), in the presence or the absence of thymidine as indicated. Growth was assessed after 2 days.
S phase arrest and checkpoint activation after thymidine depletion
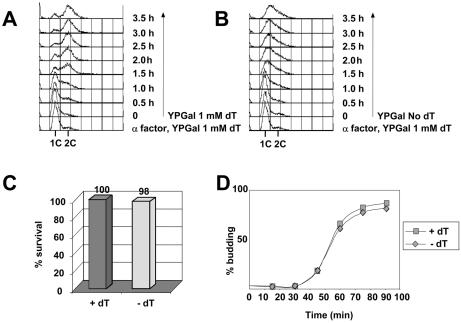
The availability of a strain whose growth is dependent upon exogenous thymidine allowed us to examine the consequences of depleting a single nucleoside. We first examined S phase progression in the absence of thymidine using flow cytometry. Cells pre-grown in YPGal containing thymidine were arrested in G1 phase with the α factor mating pheromone. They were then released into YPGal medium either in the presence (Fig. 2A) or absence (Fig. 2B) of thymidine. Nocodazole was added to prevent progression through mitosis. In the presence of thymidine, S phase started between 30 and 60 min after release and, by 90 min, most of the cells had reached a 2C content indicating that they had finished DNA replication. In the absence of thymidine, S phase began at the same time, but progression was blocked as cells accumulated with a DNA content between 1C and 2C, indicating that removing thymidine from the medium prevents completion of the first S phase after release from a G1 arrest. Figure 2D shows that bud emergence, which is a Start-dependent process independent of DNA replication, was unaffected by thymidine depletion. Figure 2C shows that the viability of the cells after 4 h in the absence of thymidine was 98% of the control in the presence of the nucleoside, indicating that the arrest in S phase due to thymidine depletion is reversible.
Figure 2.
Thymidine depletion provokes an S phase arrest. (A and B) YLVA10 cells were grown in YPGal containing 100 µM thymidine until they reached a density of 5 × 107 cells/ml. They were then arrested in G1 with α factor and released in YPGal in the presence (A) or the absence (B) of 1 mM thymidine. The DNA content of yeast cells was measured by flow cytometry. (C) After 2.5 h (A and B), equal volumes of cells were plated onto YPGal plates containing 100 µM thymidine. Viability was determined by comparing the number of colonies after release from α factor to the number of colonies in the α factor arrest. (D) Budding index of cells in S phase after release from G1 in the presence (+dT) or in the absence (–dT) of 100 µM thymidine.
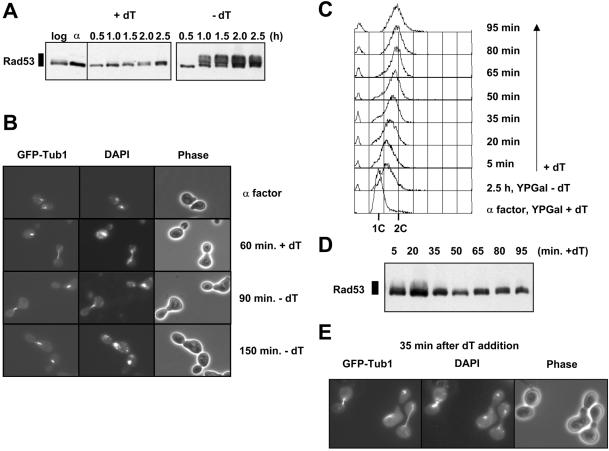
The S phase checkpoint is activated when all four dNTPs are depleted by inhibition of ribonucleotide reductase with hydroxyurea leading to hyperphosphorylation of the checkpoint kinase Rad53 (23). We asked whether the depletion of a single nucleotide was sufficient to cause Rad53 hyperphosphorylation. In the experiment described above, extracts from samples were analysed by immunoblotting using anti-Rad53 antibody (24). Rad53 was not hyperphosphorylated in an asynchronous culture or when cells were blocked in G1 with α factor. We often observe a small amount of Rad53 phosphorylation in asynchronous cells, which may be a consequence of imbalanced dNTP pools. After release from α factor in the presence of thymidine, Rad53 was also not phosphorylated (Fig. 3A), indicating that S phase probably occurs normally in this strain, in accordance with the flow cytometry (Fig. 2A). When cells were released from α factor in the absence of thymidine, however, Rad53 became hyperphosphorylated by 60 min as indicated by the appearance of slower migrating forms. The hyperphosphorylation of Rad53 coincides with the start of S phase (Fig. 2A and B, 60 min) and these slower migrating forms of Rad53 remained whilst exogenous thymidine was absent. At later times the slowest migrating forms of Rad53 began to disappear, perhaps because of checkpoint adaptation (25). Thus, thymidine depletion is rapidly sensed by the cells entering S phase and Rad53 becomes phosphorylated, indicating that the S phase checkpoint is activated under these conditions.
Figure 3.
Reversible S phase checkpoint activation after thymidine depletion. (A) Hyper-phosphorylation of Rad53 after thymidine depletion. As described in Figure 2A and B, YLVA10 cells were grown in YPGal containing 100 µM thymidine and were arrested in G1 with α factor. After release in YPGal in the presence (+dT) or the absence (–dT) of 100 µM thymidine, samples were taken and processed for immunoblotting to detect Rad53 with a polyclonal antibody. The DNA content of yeast cells was measured by flow cytometry. (B) Thymidine depletion inhibits microtubule elongation. Samples were collected at indicated time points and processed for observation under the microscope. Spindle microtubules (GFP-Tub1) are shown in the left panels. DNA, visualised by DAPI staining, is shown in the central panels. Phase contrast images of the cells are shown in the right panels. (C–E) S phase arrest due to thymidine depletion is reversible. Cells were arrested in G1 with α factor, and released into YPGal without thymidine. After 2.5 h, 100 µM thymidine was added to the culture, and samples were taken every 15 min and processed for FACS analysis (C), immunoblot analysis using anti-Rad53 antibody (D) and GFP-Tubulin (E).
S phase checkpoint activation inhibits the metaphase to anaphase transition thus preventing mitotic microtubule elongation. In order to look at whether microtubule elongation is inhibited in this strain in the absence of thymidine, we expressed a protein fusion of GFP and α-tubulin (Tub1) (15). Cells were blocked in α factor and released into YPGal, either supplemented with thymidine or containing no thymidine. In the presence of thymidine, microtubules elongated normally from 60 min after release onwards, and the nucleus divided accordingly as shown by DAPI staining (Fig. 3B). When S phase was arrested by thymidine depletion, tubulin was detected as a short, thick bar of fluorescence, indicating that cells had not yet elongated their microtubules 1.5 and 2.5 h after release. The survival of cells in the temporary absence of thymidine (Fig. 2C) suggested that the S phase arrest is reversible. Figure 3C shows that cells complete S phase 20–35 min after re-addition of thymidine. Rad53 became dephosphorylated and mitotic spindles elongated with similar kinetics (Fig. 3D and E). Taken together, these results show that thymidine depletion from the medium provokes checkpoint activation. This activation is reversible, and is rapidly relieved following thymidine addition.
Incorporation of BrdU and use in pulse–chase experiments
We are interested in examining the effects of various nucleoside analogues on DNA replication and checkpoint control. We reasoned that expression of a nucleoside kinase with broader substrate specificity might be useful for this. In contrast to all known deoxyribonucleoside kinases, a single highly efficient deoxyribonucleoside kinase from D.melanogaster (DmdNK) is able to phosphorylate all precursor nucleosides for DNA synthesis (11,26). Moreover, a series of mutants in DmdNK have been isolated which have relaxed specificities for a number of clinically important nucleoside analogues (27). Therefore, we constructed a strain expressing wild type DmdNK, together with hENT1 in a CDC21 null background. Similar to the HSV-TK-expressing strains, this strain requires exogenous thymidine for growth (data not shown).
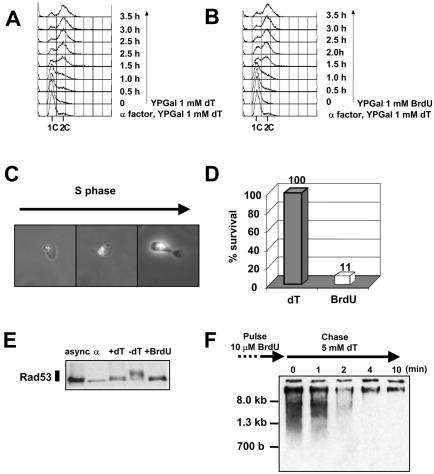
BrdU is a thymidine analogue in which the methyl group on the thymine base has been substituted with bromine. It is efficiently incorporated into the DNA of mammalian cells, but this incorporation is inefficient in yeast. BrdU incorporation would be useful because fully BrdU-substituted DNA is denser than normal DNA and, therefore can be used in density substitution experiments. Also, antibodies to BrdU can be used to monitor DNA synthesis. To look at S phase in the presence of BrdU in this strain, cells were blocked with α factor and released in the presence of either BrdU or thymidine. Cells were kept in the dark to prevent damage of BrdU-substituted DNA. Samples were analysed by flow cytometry. In the presence of BrdU, S phase starts and finishes with very similar timing compared with S phase in the presence of thymidine (Fig. 4A and B), suggesting that BrdU is taken up and incorporated efficiently in the DNA in this strain. To confirm the incorporation of BrdU into DNA we used anti-BrdU antibodies to detect BrdU-labelled DNA by immunofluorescence at various stages of S phase, monitored by bud growth (Fig. 4C). Early stages of S phase show a discrete punctate pattern, whereas later stages of S phase show a more intense, continuous fluorescence.
Figure 4.
Efficient BrdU incorporation into yeast DNA. (A and B) YLVE4 cells were arrested in G1 with α factor, and released into YPGal in the presence of 1 mM dT (A) or the presence of 1 mM BrdU (B). The DNA content of yeast cells was measured by flow cytometry. (C) BrdU incorporated into DNA during S phase was detected by immunofluorescence using an anti-BrdU antibody (see Materials and Methods). Phase contrast image was superimposed to UV light image so that cell morphology is visible. (D) After 2.5 h viability was determined as in Figure 2C. (E) Rad53 is not phosphorylated in the presence of BrdU. Samples were taken 2.5 h after release from α factor in YPGal + Nocodazole + 1 mM dT, YPGal + Nocodazole or YPGal+ Nocodazole + 1 mM BrdU, and processed for immunoblotting to detect Rad53. (F) Immunodetection of BrdU-pulsed DNA. DNA was run on a 0.8% alkaline agarose gel and transferred onto nylon membrane, before detection with an anti-BrdU antibody (see Materials and Methods).
Cells that had gone through a single S phase in the presence of BrdU had reduced viability compared to the control cells in the presence of thymidine (Fig. 4D). In accordance with this result, cells were unable to grow on a YPGal plate containing BrdU (data not shown). As shown in Figure 4E, this loss in viability is not accompanied by Rad53 hyperphosphorylation. Thus, BrdU incorporation into DNA does not activate the intra-S phase checkpoint in a single cell cycle, suggesting that it does not cause detectable DNA damage. The reason for the lethality is not known. Perhaps some key regulatory protein(s) like transcription factors bind poorly to fully BrdU-substituted DNA.
We tested whether this strain was suitable for pulse–chase experiments using BrdU. Asynchronous cells were grown in YPGal medium containing thymidine. Thymidine was then removed from the medium, and replaced with BrdU (10 µM). After a 10-min pulse, BrdU was chased with thymidine (5 mM) and samples were taken for further analysis. DNA was purified and run on a denaturing agarose gel, before being transferred onto a nylon membrane. Immunodetection was performed with an anti-BrdU antibody (Fig. 4F). After a 10 min pulse, single stranded DNA was detected not only at high molecular weight, but also as a smear extending down to ∼700 bp. Within 4 min after thymidine addition (chase) no low molecular weight signal was seen indicating that all of the incorporated BrdU had been chased into the high molecular weight fraction. The reduced signal at later times (e.g. 10 min) is reproducible and is due to less efficient transfer of high molecular weight DNA.
In summary, we have constructed strains that can efficiently incorporate exogenous thymidine and thymidine analogues into DNA. The pulse–chase experiment suggests that dNTP pools can be rapidly changed in this strain. We anticipate that these strains will have several uses. They will be useful for examining events like chromatin assembly on nascent DNA using pulse–chase protocols. The efficient incorporation of BrdU into DNA should be useful for density transfer experiments obviating the requirement for heavy isotopes. Variants of DmdNK exist that can incorporate a wide variety of clinically relevant nucleoside analogues into DNA. Use of these variants should allow detailed examination of the effects of nucleoside analogues on DNA replication progression and checkpoint activation. Mutant deoxyribonucleoside kinases are becoming increasingly popular as suicide genes in gene therapy. These strains should also be useful to isolate new kinase mutant-analogue pairs with the ability to sensitise yeast toward the analogue and, potentially, to use in gene therapy to sensitise target human cells.
Acknowledgments
ACKNOWLEDGEMENTS
We are very grateful to Carol Cass, University of Alberta, for advice and plasmids. We also thank Anne Early for critical reading and editing of this manuscript. L.V. was also supported by Inserm (Institut National de la Santé et de la Recherche Médicale). This work was supported by Cancer Research UK.
REFERENCES
- 1.Bell S.P. and Dutta,A. (2002) DNA replication in eukaryotic cells. Annu. Rev. Biochem., 71, 333–374. [DOI] [PubMed] [Google Scholar]
- 2.McNeil J.B. and Friesen,J.D. (1981) Expression of the Herpes simplex virus thymidine kinase gene in Saccharomyces cerevisiae. Mol. Gen. Genet., 184, 386–393. [DOI] [PubMed] [Google Scholar]
- 3.Lengronne A., Pasero,P., Bensimon,A. and Schwob,E. (2001) Monitoring S phase progression globally and locally using BrdU incorporation in TK(+) yeast strains. Nucleic Acids Res., 29, 1433–1442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dahmann C., Diffley,J.F.X. and Nasmyth,K.A. (1995) S-phase-promoting cyclin-dependent kinases prevent re-replication by inhibiting the transition of origins to a pre-replicative state. Curr. Biol., 5, 1257–1269. [DOI] [PubMed] [Google Scholar]
- 5.Sclafani R.A. and Fangman,W.L. (1986) Thymidine utilization by tut mutants and facile cloning of mutant alleles by plasmid conversion in S.cerevisiae. Genetics, 114, 753–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lemontt J.F., Fugit,D.R. and MacKay,V.L. (1980) Pleiotropic mutations at the TUP1 locus that affect the expression of mating-type-dependent functions in Saccharomyces cerevisiae. Genetics, 94, 899–920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.DeRisi J.L., Iyer,V.R. and Brown,P.O. (1997) Exploring the metabolic and genetic control of gene expression on a genomic scale. Science, 278, 680–686. [DOI] [PubMed] [Google Scholar]
- 8.Cass C.E., Young,J.D., Baldwin,S.A., Cabrita,M.A., Graham,K.A., Griffiths,M., Jennings,L.L., Mackey,J.R., Ng,A.M., Ritzel,M.W. et al. (1999) Nucleoside transporters of mammalian cells. Pharm. Biotechnol., 12, 313–352. [DOI] [PubMed] [Google Scholar]
- 9.Vickers M.F., Mani,R.S., Sundaram,M., Hogue,D.L., Young,J.D., Baldwin,S.A. and Cass,C.E. (1999) Functional production and reconstitution of the human equilibrative nucleoside transporter (hENT1) in Saccharomyces cerevisiae. Interaction of inhibitors of nucleoside transport with recombinant hENT1 and a glycosylation-defective derivative (hENT1/N48Q). Biochem. J., 339, 21–32. [PMC free article] [PubMed] [Google Scholar]
- 10.Griffiths M., Beaumont,N., Yao,S.Y., Sundaram,M., Boumah,C.E., Davies,A., Kwong,F.Y., Coe,I., Cass,C.E., Young,J.D. et al. (1997) Cloning of a human nucleoside transporter implicated in the cellular uptake of adenosine and chemotherapeutic drugs. Nature Med., 3, 89–93. [DOI] [PubMed] [Google Scholar]
- 11.Munch-Petersen B., Piskur,J. and Sondergaard,L. (1998) Four deoxynucleoside kinase activities from Drosophila melanogaster are contained within a single monomeric enzyme, a new multifunctional deoxynucleoside kinase. J. Biol. Chem., 273, 3926–3931. [DOI] [PubMed] [Google Scholar]
- 12.Knecht W., Munch-Petersen,B. and Piskur,J. (2000) Polyclonal antibodies against the ultrafast multisubstrate deoxyribonucleoside kinase from Drosophila melanogaster. Adv. Exp. Med. Biol., 486, 263–266. [DOI] [PubMed] [Google Scholar]
- 13.Taylor G.R., Barclay,B.J., Storms,R.K., Friesen,J.D. and Haynes,R.H. (1982) Isolation of the thymidylate synthetase gene (TMP1) by complementation in Saccharomyces cerevisiae. Mol. Cell. Biol., 2, 437–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Carreras C.W. and Santi,D.V. (1995) The catalytic mechanism and structure of thymidylate synthase. Annu. Rev. Biochem., 64, 721–762. [DOI] [PubMed] [Google Scholar]
- 15.Straight A.F., Marshall,W.F., Sedat,J.W. and Murray,A.W. (1997) Mitosis in living budding yeast: anaphase A but no metaphase plate. Science, 277, 574–578. [DOI] [PubMed] [Google Scholar]
- 16.Sambrook J., Fritsch,E.F. and Maniatis,T. (1989) Molecular Cloning: A Laboratory Manual, 2nd Edn. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 17.Santocanale C. and Diffley,J.F.X. (1998) A Mec1- and Rad53-dependent checkpoint controls late-firing origins of DNA replication. Nature, 395, 615–618. [DOI] [PubMed] [Google Scholar]
- 18.Neff M.W. and Burke,D.J. (1991) Random segregation of chromatids at mitosis in Saccharomyces cerevisiae. Genetics, 127, 463–473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Santocanale C., Neecke,H., Longhese,M.P., Lucchini,G. and Plevani,P. (1995) Mutations in the gene encoding the 34 kDa subunit of yeast replication protein A cause defective S phase progression. J. Mol. Biol., 254, 595–607. [DOI] [PubMed] [Google Scholar]
- 20.Lin H.B., Falchetto,R., Mosca,P.J., Shabanowitz,J., Hunt,D.F. and Hamlin,J.L. (1996) Mimosine targets serine hydroxymethyltransferase. J. Biol. Chem., 271, 2548–2556. [DOI] [PubMed] [Google Scholar]
- 21.Schweitzer B.I., Dicker,A.P. and Bertino,J.R. (1990) Dihydrofolate reductase as a therapeutic target. FASEB J., 4, 2441–2452. [DOI] [PubMed] [Google Scholar]
- 22.Anand N. (1996) In Wolff,M.E. (ed.), Burger’s Medicinal Chemistry and Drug Discovery. John Wiley & Sons, New York, NY, pp. 527–573. [Google Scholar]
- 23.Sun Z., Fay,D.S., Marini,F., Foiani,M. and Stern,D.F. (1996) Spk1/Rad53 is regulated by Mec1-dependent protein phosphorylation in DNA replication and damage checkpoint pathways. Genes Dev., 10, 395–406. [DOI] [PubMed] [Google Scholar]
- 24.Tercero J.A., Longhese,M.P. and Diffley,J.F.X. (2003) A central role for DNA replication forks in checkpoint activation and response. Mol. Cell, 11, 1323–1336. [DOI] [PubMed] [Google Scholar]
- 25.Toczyski D.P., Galgoczy,D.J. and Hartwell,L.H. (1997) CDC5 and CKII control adaptation to the yeast DNA damage checkpoint. Cell, 90, 1097–1106. [DOI] [PubMed] [Google Scholar]
- 26.Munch-Petersen B., Knecht,W., Lenz,C., Sondergaard,L. and Piskur,J. (2000) Functional expression of a multisubstrate deoxyribonucleoside kinase from Drosophila melanogaster and its C-terminal deletion mutants. J. Biol. Chem., 275, 6673–6679. [DOI] [PubMed] [Google Scholar]
- 27.Knecht W., Munch-Petersen,B. and Piskur,J. (2000) Identification of residues involved in the specificity and regulation of the highly efficient multisubstrate deoxyribonucleoside kinase from Drosophila melanogaster. J. Mol. Biol., 301, 827–837. [DOI] [PubMed] [Google Scholar]