Abstract
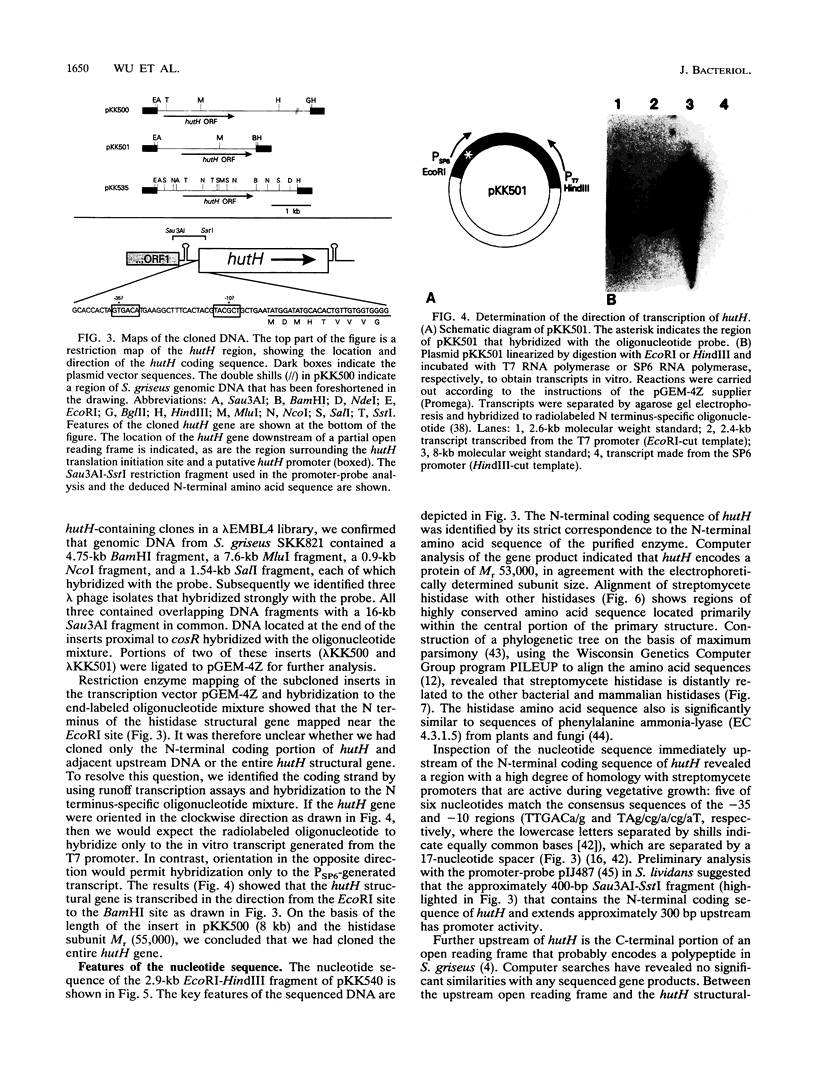
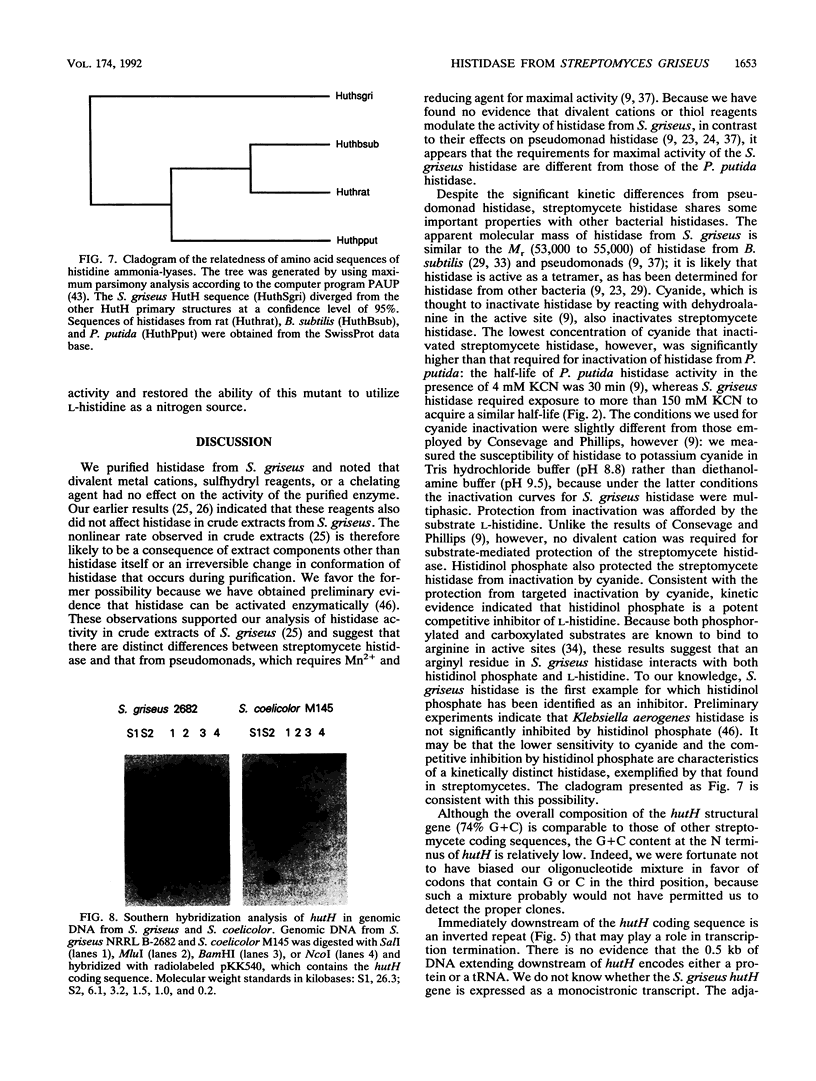
Histidine ammonia-lyase (histidase) was purified to homogeneity from vegetative mycelia of Streptomyces griseus. The enzyme was specific for L-histidine and showed no activity against the substrate analog, D-histidine. Histidinol phosphate was a potent competitive inhibitor. Histidase displayed saturation kinetics with no detectable sigmoidal response. Neither thiol reagents nor a variety of divalent cations had any effect on the activity of the purified enzyme. High concentrations of potassium cyanide inactivated histidase in the absence of its substrate or histidinol phosphate, suggesting that, as in other histidases, dehydroalanine plays an important role in catalysis. The N-terminal amino acid sequence of histidase was used to construct a mixed oligonucleotide probe to identify and clone the histidase structural gene, hutH, from genomic DNA of the wild-type strain of S. griseus. The cloned DNA restored the ability of a histidase structural gene mutant to grow on L-histidine as the sole nitrogen source. The deduced amino acid sequence of hutH shows significant relatedness with histidase from bacteria and a mammal as well as phenylalanine ammonia-lyase from plants and fungi.
Full text
PDF

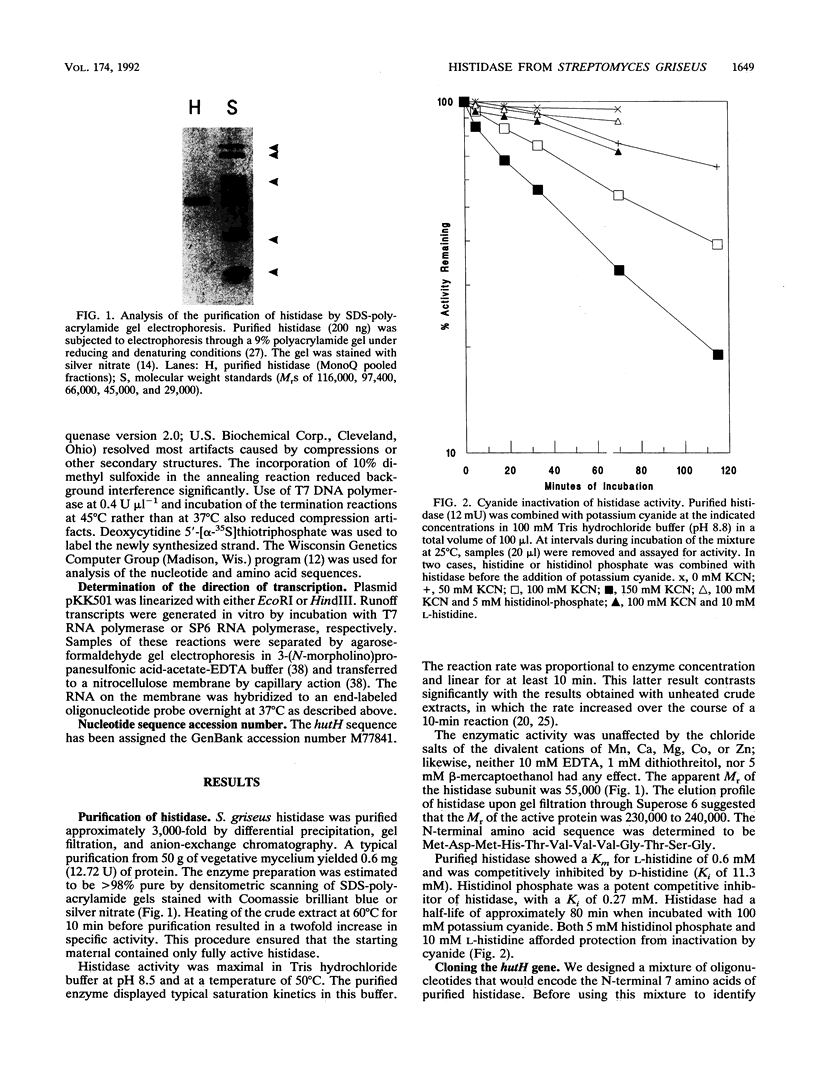



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Allison S. L., Phillips A. T. Nucleotide sequence of the gene encoding the repressor for the histidine utilization genes of Pseudomonas putida. J Bacteriol. 1990 Sep;172(9):5470–5476. doi: 10.1128/jb.172.9.5470-5476.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babcock M. J., Kendrick K. E. Cloning of DNA involved in sporulation of Streptomyces griseus. J Bacteriol. 1988 Jun;170(6):2802–2808. doi: 10.1128/jb.170.6.2802-2808.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babcock M. J., Kendrick K. E. Transcriptional and translational features of a sporulation gene of Streptomyces griseus. Gene. 1990 Oct 30;95(1):57–63. doi: 10.1016/0378-1119(90)90413-l. [DOI] [PubMed] [Google Scholar]
- Bibb M. J., Findlay P. R., Johnson M. W. The relationship between base composition and codon usage in bacterial genes and its use for the simple and reliable identification of protein-coding sequences. Gene. 1984 Oct;30(1-3):157–166. doi: 10.1016/0378-1119(84)90116-1. [DOI] [PubMed] [Google Scholar]
- Blumenberg M., Magasanik B. Physical maps of Klebsiella aerogenes and Salmonella typhimurium hut genes. J Bacteriol. 1981 Jan;145(1):664–667. doi: 10.1128/jb.145.1.664-667.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boylan S. A., Bender R. A. Genetic and physical maps of Klebsiella aerogenes genes for histidine utilization (hut). Mol Gen Genet. 1984;193(1):99–103. doi: 10.1007/BF00327421. [DOI] [PubMed] [Google Scholar]
- Brand L. M., Harper A. E. Histidine ammonia-lyase from rat liver. Purification, properties, and inhibition by substrate analogues. Biochemistry. 1976 May 4;15(9):1814–1821. doi: 10.1021/bi00654a005. [DOI] [PubMed] [Google Scholar]
- Chasin L. A., Magasanik B. Induction and repression of the histidine-degrading enzymes of Bacillus subtilis. J Biol Chem. 1968 Oct 10;243(19):5165–5178. [PubMed] [Google Scholar]
- Consevage M. W., Phillips A. T. Presence and quantity of dehydroalanine in histidine ammonia-lyase from Pseudomonas putida. Biochemistry. 1985 Jan 15;24(2):301–308. doi: 10.1021/bi00323a010. [DOI] [PubMed] [Google Scholar]
- Consevage M. W., Phillips A. T. Sequence analysis of the hutH gene encoding histidine ammonia-lyase in Pseudomonas putida. J Bacteriol. 1990 May;172(5):2224–2229. doi: 10.1128/jb.172.5.2224-2229.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coote J. G., Hassall H. The control of the enzymes degrading histidine and related imidazolyl derivates in Pseudomonas testosteroni. Biochem J. 1973 Mar;132(3):423–433. doi: 10.1042/bj1320423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devereux J., Haeberli P., Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 1):387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehresmann B., Imbault P., Weil J. H. Spectrophotometric determination of protein concentration in cell extracts containing tRNA's and rRNA's. Anal Biochem. 1973 Aug;54(2):454–463. doi: 10.1016/0003-2697(73)90374-6. [DOI] [PubMed] [Google Scholar]
- Gerton G. L., Millette C. F. Stage-specific synthesis and fucosylation of plasma membrane proteins by mouse pachytene spermatocytes and round spermatids in culture. Biol Reprod. 1986 Nov;35(4):1025–1035. doi: 10.1095/biolreprod35.4.1025. [DOI] [PubMed] [Google Scholar]
- Henikoff S. Unidirectional digestion with exonuclease III creates targeted breakpoints for DNA sequencing. Gene. 1984 Jun;28(3):351–359. doi: 10.1016/0378-1119(84)90153-7. [DOI] [PubMed] [Google Scholar]
- Hu L., Phillips A. T. Organization and multiple regulation of histidine utilization genes in Pseudomonas putida. J Bacteriol. 1988 Sep;170(9):4272–4279. doi: 10.1128/jb.170.9.4272-4279.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janssen G. R., Ward J. M., Bibb M. J. Unusual transcriptional and translational features of the aminoglycoside phosphotransferase gene (aph) from Streptomyces fradiae. Genes Dev. 1989 Mar;3(3):415–429. doi: 10.1101/gad.3.3.415. [DOI] [PubMed] [Google Scholar]
- Kendrick K. E., Ensign J. C. Sporulation of Streptomyces griseus in submerged culture. J Bacteriol. 1983 Jul;155(1):357–366. doi: 10.1128/jb.155.1.357-366.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kendrick K. E., Wheelis M. L. Histidine dissimilation in Streptomyces coelicolor. J Gen Microbiol. 1982 Sep;128(9):2029–2040. doi: 10.1099/00221287-128-9-2029. [DOI] [PubMed] [Google Scholar]
- Klee C. B. Reversible polymerization of histidine ammonia lyase. The role of sulfhydryl groups in the activity and polymeric state of the enzyme. J Biol Chem. 1970 Jun;245(12):3143–3152. [PubMed] [Google Scholar]
- Kroening T. A., Kendrick K. E. Cascading regulation of histidase activity in Streptomyces griseus. J Bacteriol. 1989 Feb;171(2):1100–1105. doi: 10.1128/jb.171.2.1100-1105.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroening T. A., Kendrick K. E. In vivo regulation of histidine ammonia-lyase activity from Streptomyces griseus. J Bacteriol. 1987 Feb;169(2):823–829. doi: 10.1128/jb.169.2.823-829.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Leidigh B. J., Wheelis M. L. Genetic control of the histidine dissimilatory pathway in Pseudomonas putida. Mol Gen Genet. 1973 Feb 2;120(3):201–210. doi: 10.1007/BF00267152. [DOI] [PubMed] [Google Scholar]
- McClard R. W., Kolenbrander H. M. Resolution of temperature-dependent conformers of histidine ammonia-lyase on disc gel electrophoresis: correlation with Arrhenius discontinuities. Experientia. 1974 Jul 15;30(7):730–731. doi: 10.1007/BF01924152. [DOI] [PubMed] [Google Scholar]
- Meiss H. K., Brill W. J., Magasanik B. Genetic control of histidine degradation in Salmonella typhimurium, strain LT-2. J Biol Chem. 1969 Oct 10;244(19):5382–5391. [PubMed] [Google Scholar]
- Newell C. P., Lessie T. G. Induction of histidine-degrading enzymes in Pseudomonas aeruginosa. J Bacteriol. 1970 Oct;104(1):596–598. doi: 10.1128/jb.104.1.596-598.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oda M., Sugishita A., Furukawa K. Cloning and nucleotide sequences of histidase and regulatory genes in the Bacillus subtilis hut operon and positive regulation of the operon. J Bacteriol. 1988 Jul;170(7):3199–3205. doi: 10.1128/jb.170.7.3199-3205.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patthy L., Thész J. Origin of the selectivity of alpha-dicarbonyl reagents for arginyl residues of anion-binding sites. Eur J Biochem. 1980 Apr;105(2):387–393. doi: 10.1111/j.1432-1033.1980.tb04512.x. [DOI] [PubMed] [Google Scholar]
- Prival M. J., Magasanik B. Resistance to catabolite repression of histidase and proline oxidase during nitrogen-limited growth of Klebsiella aerogenes. J Biol Chem. 1971 Oct 25;246(20):6288–6296. [PubMed] [Google Scholar]
- Rechler M. M. The purification and characterization of L-histidine ammonia-lyse (Pseudomonas). J Biol Chem. 1969 Feb 25;244(4):551–559. [PubMed] [Google Scholar]
- Schlesinger S., Scotto P., Magasanik B. Exogenous and endogenous induction of the histidine-degrading enzymes in Aerobacter aerogenes. J Biol Chem. 1965 Nov;240(11):4331–4337. [PubMed] [Google Scholar]
- Schwacha A., Bender R. A. Nucleotide sequence of the gene encoding the repressor for the histidine utilization genes of Klebsiella aerogenes. J Bacteriol. 1990 Sep;172(9):5477–5481. doi: 10.1128/jb.172.9.5477-5481.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Taylor R. G., Lambert M. A., Sexsmith E., Sadler S. J., Ray P. N., Mahuran D. J., McInnes R. R. Cloning and expression of rat histidase. Homology to two bacterial histidases and four phenylalanine ammonia-lyases. J Biol Chem. 1990 Oct 25;265(30):18192–18199. [PubMed] [Google Scholar]
- Ward J. M., Janssen G. R., Kieser T., Bibb M. J., Buttner M. J., Bibb M. J. Construction and characterisation of a series of multi-copy promoter-probe plasmid vectors for Streptomyces using the aminoglycoside phosphotransferase gene from Tn5 as indicator. Mol Gen Genet. 1986 Jun;203(3):468–478. doi: 10.1007/BF00422072. [DOI] [PubMed] [Google Scholar]