Abstract
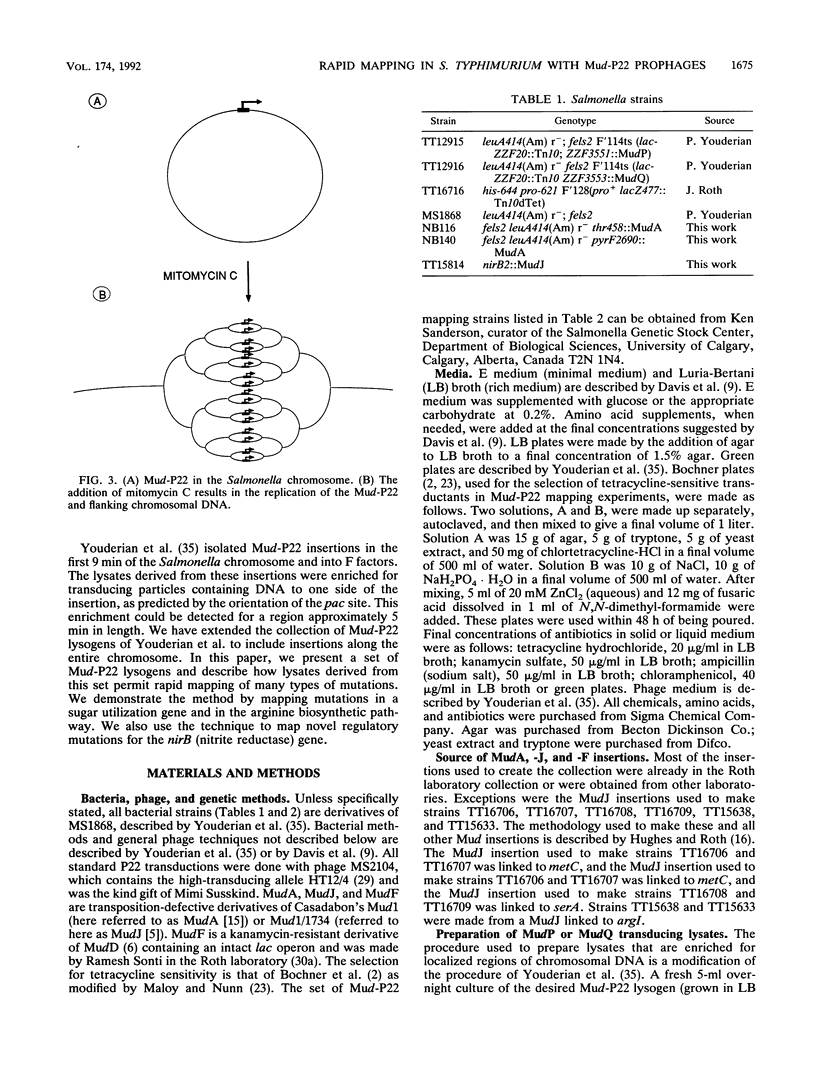
A new method for mapping mutations in the Salmonella typhimurium chromosome is described and applied to the localization of novel regulatory mutations affecting expression of the nirB (nitrite reductase) gene. The mapping technique is also illustrated by the mapping of mutations in genes affecting carbohydrate catabolism and biosynthetic pathways. The new mapping method involves use of the hybrid phage MudP and MudQ (together referred to as Mud-P22), originally constructed by Youderian et al. (Genetics 118:581-592, 1988). This report describes a set of Mud-P22 lysogens, each member of the set containing a different Mud-P22 insertion. The insertions are scattered along the entire Salmonella genome. These lysogens, when induced by mitomycin C, generate transducing lysates that are enriched (45- to 1,400-fold over the background, generalized transducing particle population) for transducing particles containing bacterial DNA that flanks one side of the insertion. We demonstrate that within the set of lysogens there can be found at least one Mud-P22 insertion that enriches for any particular region of the Salmonella chromosome and that, therefore, all regions of the chromosome are discretely enriched and represented by the collection as a whole. We describe a technique that allows the rapid and facile determination of which lysate contains enriched sequences for the repair of a mutant locus, thereby allowing the determination of the map position of the locus. This technique is applicable to those mutations for which the wild-type allele is selectable. We also describe a procedure whereby any Tn10 insertion can be mapped by selecting for the loss of Tetr.
Full text
PDF







Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Adams M. B., Hayden M., Casjens S. On the sequential packaging of bacteriophage P22 DNA. J Virol. 1983 May;46(2):673–677. doi: 10.1128/jvi.46.2.673-677.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bochner B. R., Huang H. C., Schieven G. L., Ames B. N. Positive selection for loss of tetracycline resistance. J Bacteriol. 1980 Aug;143(2):926–933. doi: 10.1128/jb.143.2.926-933.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casjens S., Hayden M. Analysis in vivo of the bacteriophage P22 headful nuclease. J Mol Biol. 1988 Feb 5;199(3):467–474. doi: 10.1016/0022-2836(88)90618-3. [DOI] [PubMed] [Google Scholar]
- Casjens S., Huang W. M., Hayden M., Parr R. Initiation of bacteriophage P22 DNA packaging series. Analysis of a mutant that alters the DNA target specificity of the packaging apparatus. J Mol Biol. 1987 Apr 5;194(3):411–422. doi: 10.1016/0022-2836(87)90671-1. [DOI] [PubMed] [Google Scholar]
- Castilho B. A., Olfson P., Casadaban M. J. Plasmid insertion mutagenesis and lac gene fusion with mini-mu bacteriophage transposons. J Bacteriol. 1984 May;158(2):488–495. doi: 10.1128/jb.158.2.488-495.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaconas G., de Bruijn F. J., Casadaban M. J., Lupski J. R., Kwoh T. J., Harshey R. M., DuBow M. S., Bukhari A. I. In vitro and in vivo manipulations of bacteriophage Mu DNA: cloning of Mu ends and construction of mini-Mu's carrying selectable markers. Gene. 1981 Jan-Feb;13(1):37–46. doi: 10.1016/0378-1119(81)90041-x. [DOI] [PubMed] [Google Scholar]
- Chelala C. A., Margolin P. Effects of deletions on cotransduction linkage in Salmonella typhimurium: evidence that bacterial chromosome deletions affect the formation of transducing DNA fragments. Mol Gen Genet. 1974;131(2):97–112. doi: 10.1007/BF00266146. [DOI] [PubMed] [Google Scholar]
- Chumley F. G., Menzel R., Roth J. R. Hfr formation directed by tn10. Genetics. 1979 Apr;91(4):639–655. doi: 10.1093/genetics/91.4.639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebel-Tsipis J., Botstein D., Fox M. S. Generalized transduction by phage P22 in Salmonella typhimurium. I. Molecular origin of transducing DNA. J Mol Biol. 1972 Nov 14;71(2):433–448. doi: 10.1016/0022-2836(72)90361-0. [DOI] [PubMed] [Google Scholar]
- Ebel-Tsipis J., Fox M. S., Botstein D. Generalized transduction by bacteriophage P22 in Salmonella typhimurium. II. Mechanism of integration of transducing DNA. J Mol Biol. 1972 Nov 14;71(2):449–469. doi: 10.1016/0022-2836(72)90362-2. [DOI] [PubMed] [Google Scholar]
- Hughes K. T., Roth J. R. Conditionally transposition-defective derivative of Mu d1(Amp Lac). J Bacteriol. 1984 Jul;159(1):130–137. doi: 10.1128/jb.159.1.130-137.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes K. T., Roth J. R. Transitory cis complementation: a method for providing transposition functions to defective transposons. Genetics. 1988 May;119(1):9–12. doi: 10.1093/genetics/119.1.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson E. N., Jackson D. A., Deans R. J. EcoRI analysis of bacteriophage P22 DNA packaging. J Mol Biol. 1978 Jan 25;118(3):365–388. doi: 10.1016/0022-2836(78)90234-6. [DOI] [PubMed] [Google Scholar]
- Jackson E. N., Laski F., Andres C. Bacteriophage P22 mutants that alter the specificity of DNA packaging. J Mol Biol. 1982 Feb 5;154(4):551–563. doi: 10.1016/s0022-2836(82)80014-4. [DOI] [PubMed] [Google Scholar]
- Kukral A. M., Strauch K. L., Maurer R. A., Miller C. G. Genetic analysis in Salmonella typhimurium with a small collection of randomly spaced insertions of transposon Tn10 delta 16 delta 17. J Bacteriol. 1987 May;169(5):1787–1793. doi: 10.1128/jb.169.5.1787-1793.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LEDERBERG J., LEDERBERG E. M., ZINDER N. D., LIVELY E. R. Recombination analysis of bacterial heredity. Cold Spring Harb Symp Quant Biol. 1951;16:413–443. doi: 10.1101/sqb.1951.016.01.030. [DOI] [PubMed] [Google Scholar]
- Laski F., Jackson E. N. Maturation cleavage of bacteriophage P22 DNA in the absence of DNA packaging. J Mol Biol. 1982 Feb 5;154(4):565–579. doi: 10.1016/s0022-2836(82)80015-6. [DOI] [PubMed] [Google Scholar]
- Liu S. L., Sanderson K. E. A physical map of the Salmonella typhimurium LT2 genome made by using XbaI analysis. J Bacteriol. 1992 Mar;174(5):1662–1672. doi: 10.1128/jb.174.5.1662-1672.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maloy S. R., Nunn W. D. Selection for loss of tetracycline resistance by Escherichia coli. J Bacteriol. 1981 Feb;145(2):1110–1111. doi: 10.1128/jb.145.2.1110-1111.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearce U., Stocker B. A. Variation in composition of chromosome fragments transduced by phage P22. Virology. 1965 Nov;27(3):290–296. doi: 10.1016/0042-6822(65)90108-x. [DOI] [PubMed] [Google Scholar]
- Raj A. S., Raj A. Y., Schmieger H. Phage genes involved in the formation generalized transducing particles in Salmonella--Phage P22. Mol Gen Genet. 1974;135(2):175–184. doi: 10.1007/BF00264784. [DOI] [PubMed] [Google Scholar]
- Roth J. R., Hartman P. E. Heterogeneity in P22 transducing particles. Virology. 1965 Nov;27(3):297–307. doi: 10.1016/0042-6822(65)90109-1. [DOI] [PubMed] [Google Scholar]
- Sanderson K. E., Roth J. R. Linkage map of Salmonella typhimurium, edition VII. Microbiol Rev. 1988 Dec;52(4):485–532. doi: 10.1128/mr.52.4.485-532.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmieger H. Phage P22-mutants with increased or decreased transduction abilities. Mol Gen Genet. 1972;119(1):75–88. doi: 10.1007/BF00270447. [DOI] [PubMed] [Google Scholar]
- Smith C. M., Koch W. H., Franklin S. B., Foster P. L., Cebula T. A., Eisenstadt E. Sequence analysis and mapping of the Salmonella typhimurium LT2 umuDC operon. J Bacteriol. 1990 Sep;172(9):4964–4978. doi: 10.1128/jb.172.9.4964-4978.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Susskind M. M., Botstein D. Molecular genetics of bacteriophage P22. Microbiol Rev. 1978 Jun;42(2):385–413. doi: 10.1128/mr.42.2.385-413.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel W., Schmieger H. Selection of bacterial pac sites recognized by Salmonella phage P22. Mol Gen Genet. 1986 Dec;205(3):563–567. doi: 10.1007/BF00338100. [DOI] [PubMed] [Google Scholar]
- Weaver S., Levine M. Replication in situ and DNA encapsulation following induction of an excision-defective lysogen of Salmonella bacteriophage P22. J Mol Biol. 1978 Jan 25;118(3):389–411. doi: 10.1016/0022-2836(78)90235-8. [DOI] [PubMed] [Google Scholar]
- Wong K. K., McClelland M. A BlnI restriction map of the Salmonella typhimurium LT2 genome. J Bacteriol. 1992 Mar;174(5):1656–1661. doi: 10.1128/jb.174.5.1656-1661.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youderian P., Sugiono P., Brewer K. L., Higgins N. P., Elliott T. Packaging specific segments of the Salmonella chromosome with locked-in Mud-P22 prophages. Genetics. 1988 Apr;118(4):581–592. doi: 10.1093/genetics/118.4.581. [DOI] [PMC free article] [PubMed] [Google Scholar]






