Abstract
Phospholipases A2 (PLA2) comprise a superfamily of enzymes that hydrolyze phospholipids to a free fatty acid, e.g., arachidonate, and a 2-lysophospholipid. Dissecting their individual functions has relied in large part on pharmacological inhibitors that discriminate among PLA2. Group VIA PLA2 (iPLA2β) has a GTSTG serine lipase consensus sequence, and studies with a bromoenol lactone (BEL) suicide substrate inhibitor have been taken to suggest that iPLA2β participates in a wide variety of biological processes. Such conclusions presume inhibitor specificity. Inhibition by BEL requires its hydrolysis by and results in uncharacterized covalent modification(s) of iPLA2β. We performed mass spectrometric analyses of proteolytic digests of BEL-treated iPLA2β to identify modifications associated with loss of activity. The GTSTG active site and large flanking regions of sequence are not modified by BEL treatment, but most iPLA2β Cys residues are alkylated at various BEL concentrations to form a thioether linkage to a BEL keto acid hydrolysis product. Synthetic Cys-containing peptides are alkylated when incubated with iPLA2β and BEL, which reflects iPLA2β-catalyzed BEL hydrolysis to a diffusible bromomethyl keto acid product that reacts with distant thiols. The BEL concentration dependence of Cys651 alkylation closely parallels that of loss of iPLA2β activity. No amino acid residues other than Cys were found to be modified, suggesting that Cys alkylation is the covalent modification of iPLA2β responsible for loss of activity, and the alkylating species appears to be a diffusible hydrolysis product of BEL rather than a tethered acyl-enzyme intermediate.
Phospholipases A2 (PLA2)1 catalyze hydrolysis of the sn-2 fatty acid substituent from glycerophospholipid substrates to yield a free fatty acid, e.g., arachidonic acid, and a 2-lysophospholipid that have intrinsic mediator activities and are precursors of other mediators, including prostaglandins, thromboxanes, leukotrienes, and platelet activating factor (PAF) (1–5). Mammalian PLA2s include low molecular weight secretory PLA2 (sPLA2) that requires millimolar [Ca2+] for catalysis and affects inflammation and other processes and the PAF–acetylhydrolase PLA2 family (3). Of group IV cytosolic PLA2 (cPLA2) family members (3), cPLA2α prefers substrates with sn-2 arachidonoyl residues, associates with its substrates in membranes upon rises in cytosolic [Ca2+], and is also regulated by phosphorylation (6). There are several other members of the cPLA2 family that arise from separate genes (7–10).
The group VI PLA2 (iPLA2) enzymes (3, 4, 11–14) do not require Ca2+ for catalysis and are inhibited by a bromoenol lactone (BEL) suicide substrate that does not inhibit sPLA2 or cPLA2 at similar concentrations (15–18). Group VIA PLA2 (iPLA2β) resides in the cytoplasm of resting cells, but group VIB PLA2 (iPLA2γ) contains a peroxisomal targeting sequence and is membrane-associated (19–22). These enzymes belong to a larger class of serine lipases that are encoded by multiple genes (23, 24). The iPLA2β enzymes cloned from various species are 84–88 kDa proteins that contain a GXSXG lipase consensus sequence and eight stretches of a repetitive motif homologous to that in the protein-binding domain of ankyrin (11–13). No crystal structures of iPLA2β or other members of the group VI PLA2 family have yet been determined.
Many potential biological functions have been proposed for iPLA2β (25–39), and the facts that multiple splice variants are differentially expressed among cells and form heterooligomers with distinct properties suggest that iPLA2β gene products might have multiple functions depending on cellular context (27, 28). Among the roles proposed for iPLA2β are participation in phospholipid remodeling (26), signaling in secretion (30, 31), apoptosis (32, 33), vasomotor regulation (34, 35), transcriptional regulation (36, 37), and eicosanoid generation (38, 39).
Many cells express multiple distinct PLA2s (13, 17, 18, 40–42), and attempts to determine their individual functions have in large part relied on pharmacologic inhibitors that discriminate among PLA2. The mechanism-based iPLA2 inhibitor BEL and its enantiomers (15, 16, 34) inhibit iPLA2 at concentrations far lower than those required to inhibit sPLA2 or cPLA2 family members (14–18), and this property has been widely exploited to discern potential biological roles for iPLA2 (25–39). BEL affects more than one target (19, 23, 24, 43, 44), however, and it was first developed as an inhibitor of serine proteases (45, 46).
BEL is a substrate for the serine hydrolases chymotrypsin (45, 46) and iPLA2β (15, 16), and its inhibitory effects require its hydrolysis by and result in uncharacterized covalent modification(s) of those enzymes (15, 16, 45, 46). Understanding the detailed mechanism whereby BEL inhibits iPLA2β could permit design of more selective inhibitors, and such information might also facilitate identifying other, unsuspected enzymes that are inhibited by BEL and that could account for some effects now attributed to iPLA2β inhibition.
Some haloenol lactone suicide substrates form stable acyl adducts with active site serine residues in enzymes that cause inactivation (47). BEL is proposed to interact with the chymotrypsin active site serine to form a short-lived acyl-enzyme bromomethyl ketone intermediate, which is thought to alkylate a nearby nucleophile in the enzyme, and the acylserine linkage is then hydrolyzed (45, 46). A similar mechanism results in alkylation of cysteine thiol groups in enzymes (47), and the side chains of other suitably positioned amino acids could in principle be nucleophilic reactants with a halomethyl ketone intermediate. To characterize covalent modifications of iPLA2β that occur upon treatment with BEL, we have performed ESI/MS/MS analyses of proteolytic digests of purified, recombinant iPLA2β incubated with BEL.
Experimental Procedures
Materials
Sf9 cells and culture medium were purchased from Invitrogen (Carlsbad, CA), TALON metal affinity resin was from Clontech (Palo Alto, CA), and 1-palmitoyl-2-[14C]-linoleoyl-sn-glycero-3-phosphocholine (16:0/[14C]18:2-GPC) was from Amersham Biosciences (Piscataway, NJ). The cysteine-containing peptides PRCGVPDVA and RGPCRAFL were synthesized by the Washington University (St. Louis, MO) Protein and Nucleic Acid Laboratory. Other chemicals were purchased from Sigma Chemical (St. Louis, MO). Solvents were purchased from Fisher Chemical (St. Louis, MO). PepMap HPLC columns and precolumns were obtained from LC-Packings (San Francisco, CA).
Cloning, Expression, and Purification of Native and His-Tagged iPLA2β Proteins
Spodoptera frugiperda (Sf9) cells were cultured as described (48–50). For protein expression, cDNA encoding iPLA2β with a polyhistidine tag sequence at the C- or N-terminus (50) was cloned into the EcoRI–SalI site of pFastBac1 baculovirus shuttle vector (Invitrogen). Sf9 cell suspensions were infected by baculovirus, collected by centrifugation, and disrupted by sonication. His-tagged proteins were purified with a TALON metal affinity column, as described (51). Aliquots of protein solutions were analyzed by SDS–PAGE. Proteins were visualized by Coomassie staining or transfer to nylon membranes and immunoblotting, as described (51).
Site-Directed Mutagenesis of Cys651 to Ala651 To Yield His-Tagged iPLA2βA651 Mutant Protein
A 2.2 kb rat iPLA2β cDNA was subcloned into the vector pBluescript II SK (Stratagene) and used for mutagenesis. Substitution of Ala651 for Cys651 was performed with the QuickChange mutagenesis kit (Stratagene, La Jolla, CA). The sequence of the forward primer was 5′-CCC TCA AGT GCC TGT AAC CGC TGT AGA TGT CTT TCG TCC-3′, and the sequence of the reverse primer was 5′-GGA CGA AAG ACA TCT ACA GCG GTT ACA GGC ACT TGA GGG-3′. The fidelity of the construct was confirmed by sequencing, and the mutated cDNA was subcloned into pFast-Bac1 vector, which was used to prepare recombinant baculovirus containing the mutant construct as an insert. The His-tagged iPLA2βA651 mutant protein was expressed in Sf9 cells and purified by immobilized metal affinity chromatography, as described above.
Incubation of Recombinant His-Tagged iPLA2β or His-Tagged iPLA2βA651 with BEL, Filtration, and On-Filter Proteolytic Digestion
A solution (100 μL, 0.2 μg/μL in 200 mM imidazole and 50 mM NaCl buffer, pH 7.8) of recombinant, purified His-tagged iPLA2β or His-tagged iPLA2βA651 was incubated (10 min, 37 °C) with various concentrations of BEL (0, 0.5, 1, 2, 5, 10, or 20 μM) added in ethanol (2 μL). An aliquot (10 μL) was removed to measure iPLA2β activity, and another (40 μL) was removed to measure free cysteine content. The remainder (50 μL) was processed for MS analysis by centrifugation through a YM-50 Microcon filter (Millipore, Billerica, MA) according to the manufacturer's instructions to collect iPLA2β protein and remove BEL-containing solution. The filter was then washed three times with 25 mM NH4HCO3 to remove residual salt and BEL, and a solution (50 μL, 20 ng/μL) of sequencing grade protease was added to the filter. Proteases used included modified trypsin (Promega Corp., Madison, WI) or modified glutamic C endopeptidase (Princeton Separations, Adelphia, NJ). The filter was gently agitated to cause the protease solution to spread over its entire surface. After incubation (6–12 h, 37 °C), the digestion mixture was centrifuged. The aqueous solution was transferred to an Eppendorf tube and adjusted to pH 2 with 0.5% formic acid for further analysis by MS or LC/MS/MS.
Assay of iPLA2β or His-Tagged iPLA2βA651 Enzymatic Activity
Ca2+-independent PLA2 enzymatic activity was assayed after ethanolic injection of the substrate 1-palmitoyl-2-[14C]linoleoyl-sn-glycero-3-phosphocholine in assay buffer (40 mM Tris, pH 7.5, 5 mM EGTA) by monitoring release of [14C]linoleate, as described (52).
Spectrophotometric Quantitation of Free Thiol Content
Free thiol content of iPLA2β was determined with Ellman's reagent (DTNB) from Molecular Probes (Eugene, OR) according to the manufacturer's instructions. DTNB was dissolved in DMSO and added (final concentration of 100 mM) to the iPLA2β solution. After incubation (5 min, room temperature), thiol concentrations in the samples were determined from measurements of absorbance at 412 nm on a Titerteck Multiskan MCC/340 microplate reader (ICN Biomedicals, Aurora, OH) with reference to a standard curve prepared with varied concentrations of l-cysteine.
Mass Spectrometric Determination of Relative Free Cysteine Thiol Content
Relative free cysteine thiol content among samples was also estimated from mass spectrometric data by quantitative determination of the relative abundance of the cysteine-containing peptide in untreated preparations of iPLA2β and in BEL-treated samples using the multiple normalization method of Steen et al. (53). The unmodified peptides used for normalization were selected by relative quantitative analyses that indicated little difference in their abundance in BEL-treated vs control iPLA2β digests. Peptides from iPLA2β that were routinely observed in proteolytic digests of both BEL-treated and untreated iPLA2β and that were never observed in modified form were grouped. Each of these peptides was quantitated relative to all of the other unmodified peptides. Peptides with a relative quantitation value close to 1.0 were selected as members of the set of unmodified peptides to which the relative abundances of other peptides were compared. Members of this set of peptides included 582SSGAAPTYFRPNGR595, 538QPAELHLFR546, 596FLDGGLLANNPTLDAMTEIHEYNQDMIR623, 38VREEGQLILLQNASNR53, 396QLQDLMPVSR405, and 7LVNTLSSVTNLFSNPFR23.
MALDI/TOF Mass Spectrometry
MALDI/TOF mass spectra were acquired with a Voyager DE STR instrument (Applied Biosystems, Foster City, CA), as described (51). For peptide analyses, a saturated solution of 3,5-dimethoxy-4-hydroxycinnamic acid in 50% acetonitrile was used as the matrix. For protein analyses, the matrix solution was sinapinic acid in 0.1% formic acid. The mass spectrometer was calibrated with insulin, cytochrome c, myoglobin, and BSA in linear mode and with trypsin autolysis peptides in reflectron mode.
LC/ESI/MS/MS Analyses
As previously described (51), samples (0.2 μL) were injected into a Micromass Cap-LC liquid chromatography system (Micromass, Manchester, U.K.) and concentrated on a PepMap C18 precolumn (300 μm × 5 mm). The precolumn was then washed (3 min, 0.1% formic acid, flow rate of 30 μL/min), and the sample was eluted onto an analytical C18 column (150 mm × 17 μm) and analyzed with a solvent gradient from solution A (3% acetonitrile) to solution B (95% acetonitrile) containing 0.1% formic acid over 50 min at a flow rate reduced from 5 μL/min to 200 nL/min by stream splitting.
LC eluant was introduced into the nanoflow source of a Micromass Q-TOF Micro mass spectrometer (Micromass, Manchester, U.K.). The source temperature was 80 °C, and the cone gas flow was 50 L/h. A voltage of 3.2 kV was applied to the nanoflow probe tip, and data were acquired in positive ion mode. Survey scans were integrated over 1 s, and MS/MS scans were integrated over 3 s. Switching from survey to MS/MS scan mode was performed in a data-dependent manner. The maximum MS/MS to survey scan ratio was three. The collision energy was 28 eV. Data were processed with Masslynx 3.5 software. Multi-point calibration was performed using selected fragment ions produced by CAD of Glu-fibrinopeptide B. MS/MS spectra were processed by Masslynx software to produce a peak list file, as described (51).
Results
His-Tagged iPLA2β Protein Expressed from Its DNA in a BaculoVirus-Sf9 Cell System and Purified on Immobilized Metal Affinity Columns Exhibits iPLA2β Activity That Is Inhibited by BEL
Sf9 cells infected with baculovirus containing cDNA encoding His-tagged iPLA2β (51) were used to express the protein (Figure 1). Cytosol from these Sf9 cells was loaded onto metal affinity columns, which were then washed to remove nonadsorbed proteins. Interaction of His-tagged iPLA2β with metal ions in the column resin was then disrupted with imidazole-containing buffers to cause desorption of His-tagged iPLA2β protein. Eluant fractions were assayed for iPLA2β enzymatic activity (Figure 1B), and the proteins were analyzed by SDS–PAGE and visualized by immunoblotting with an iPLA2β antibody (Figure 1A). The activity of purified His-tagged iPLA2β was inhibited by about 80% and nearly 100% at BEL concentrations of 5 and 20 μM, respectively (Figure 1C). After purification by immobilized metal affinity chromatography, a nearly homogeneous preparation of His-tagged iPLA2β was obtained, as illustrated by Coomassie blue staining of an SDS – PAGE analysis of the purified protein (Figure 1D).
FIGURE 1.
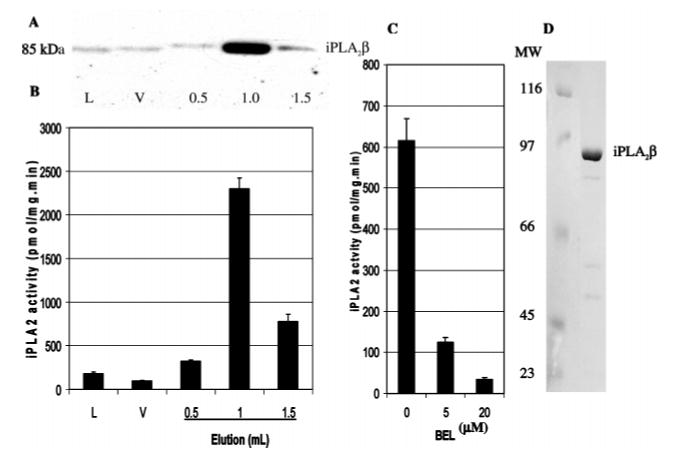
Expression of His-tagged iPLA2β in Sf9 cells and its purification on metal affinity columns. Cytosol from Sf9 cells infected with baculovirus containing DNA that encodes His-tagged iPLA2β was loaded onto a TALON immobilized metal affinity column (IMAC), which was then washed to remove weakly adherent proteins, and His-tagged iPLA2β was desorbed with buffer containing 200 mM imidazole. Proteins in aliquots of load (L), void (V), and elution fractions were analyzed by SDS–PAGE and immunoblotting with an iPLA2β antibody (panel A), and iPLA2β specific activity was also determined (panel B). Effects of 0, 5, and 20 μM BEL on the activity of purified, His-tagged iPLA2β recovered from the IMAC columns were also determined (panel C). Displayed values represent means ± SD (n = 6). The purified preparation of recombinant His-tagged iPLA2β was analyzed by SDS-PAGE with Coomassie blue staining to evaluate the purity of the protein (panel D).
Identification of Covalently Modified Amino Acid Residues in iPLA2β after Treatment with BEL
Purified, recombinant, His-tagged iPLA2β was incubated without or with varied concentrations of BEL. The BEL-containing solution was then removed by centrifugal filtration to avoid inhibition of the proteases trypsin or glutamate C, which were then used to digest iPLA2β on the filter. Proteolytic digests of BEL-treated iPLA2β were analyzed by MALDI/TOF/MS and by LC/ESI/MS/MS. Using combined data from digests with both proteases, peptides that covered over 80% of the iPLA2β sequence were identified.
By comparing MS/MS data from proteolytic digests of BEL-treated and untreated iPLA2β preparations, most cysteine residues in BEL-treated iPLA2β were found to be susceptible to covalent modification that caused peptides that contained them to exhibit a mass 254 Da greater than that for the corresponding unmodified peptides, as illustrated for peptide 238VLLLCNAR245 in Figure 2. The molecular mass of BEL is 317 Da, and the 254 Da mass increment of the adducts indicates that only a portion of the BEL molecule bound to the cysteine residues. The stability of adducts during LC/MS analyses indicates that the modification is covalent.
FIGURE 2.
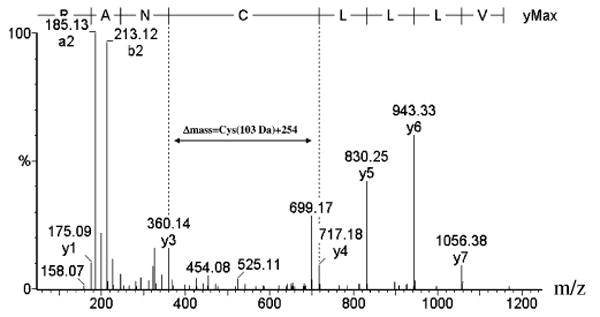
Tandem mass spectrum of the BEL-modified iPLA2β peptide 238VLLLCNAR245 produced by CAD of [M + 2H]2+(m/z 578.14). Purified His-tagged iPLA2β (10 μg) was treated with 20 μM BEL and digested with trypsin, and peptides in the digest were analyzed by LC/MS with data-dependent switching to MS/MS.
In the MS/MS spectrum of modified peptide 238VLLLCNAR245 (Figure 2), the y4–y8 ions exhibit a 254 Da shift to higher mass values, but the y1–y3 ions exhibit no shift. That indicates that Cys242 is covalently modified by a BEL-derived adduct. The strong ion at 699.17 Da in Figure 2 reflects elimination of H2O from the y4 ion, and this elimination is thought to occur from the BEL-derived moiety in the cysteine adduct. Similar losses of H2O from ions containing cysteine adducts were observed in MS/MS spectra of other modified peptides (data not shown). Evidence for modification of no amino acid residue other than cysteine was found in any of the spectra of peptides from proteolytic digests of BEL-treated iPLA2β preparations, despite the fact that peptides representing over 80% of the iPLA2β sequence were identified in such analyses.
Not every cysteine residue in the iPLA2β sequence was found to be susceptible to BEL-induced modification. For example, no peptide containing a modified Cys428 was observed, despite the fact that the unmodified peptide containing that residue was routinely identified in digests of BEL-treated iPLA2β, as illustrated in the MS/MS spectrum of the unmodified tryptic peptide containing Cys428 in Figure 3. Although Cys428 is the closest Cys residue in the linear amino acid sequence to the 463GTSTG467 catalytic center, it could well be distant from S465 in the three-dimensional space of the correctly folded enzyme.
FIGURE 3.
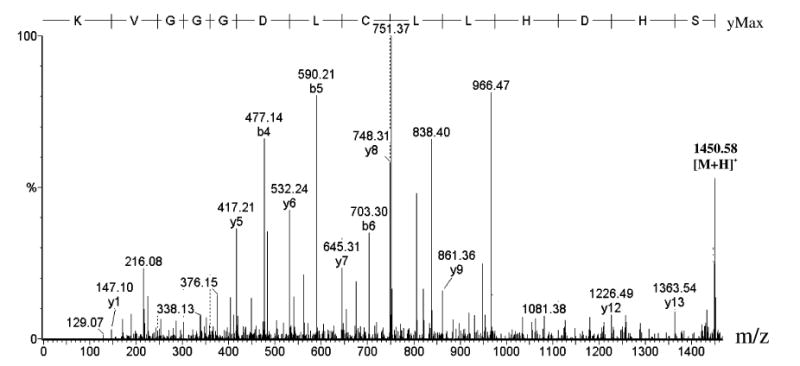
Tandem mass spectrum of the iPLA2β tryptic peptide 422SHDHLLCLDGGGVK435 produced by CAD of [M + 2H]2+ (m/z 725.79 Da).
The Serine Lipase Consensus Sequence GTSTG Is Not Covalently Modified in BEL-Treated, Catalytically Inactive iPLA2β
Another peptide that was never observed to be modified in BEL-treated iPLA2β preparations was that containing the 463GTSTG467 serine lipase consensus motif, even after treatment with 20 μM BEL under conditions that eliminate all iPLA2β activity. Stable covalent modifications of serine residues in other enzymes by similar bromoenol lactone suicide substrates are observed (47, 54), and serine residues in the catalytic centers of serine hydrolases are thought to form an acyl-enzyme intermediate with BEL (45, 46).
The interaction of BEL and iPLA2β does not result in stable modification of its active site serine residue. Figure 4 is the MS/MS spectrum of the tryptic peptide 456DLFDWVAGTSTGGI-LALAILHSK478 of iPLA2β that contains the 463GTSTG467 serine lipase consensus sequence, and this peptide was routinely observed in the tryptic digests of active, untreated iPLA2β and in those of catalytically inactive, BEL-treated iPLA2β. The spectrum contains all expected members of the y-ion series, and it also contains over half of the possible members of the b-ion series. Fragment ions y11 (1135.80 Da), y12 (1192.65 Da), y13 (1293.67 Da), y14 (1380.72 Da), y15 (1481.72 Da), and y16 (1538.7 Da) that define the amino acid sequence of the 463GTSTG467 lipase consensus are all detected with less than 0.20 Da deviation from theoretical m/z values. The routine observation of this peptide in digests of BEL-treated, catalytically inactive iPLA2β indicates that BEL does not cause stable covalent modification of S465 or other residues in the lipase consensus site or in the larger tryptic peptide that contains it.
FIGURE 4.
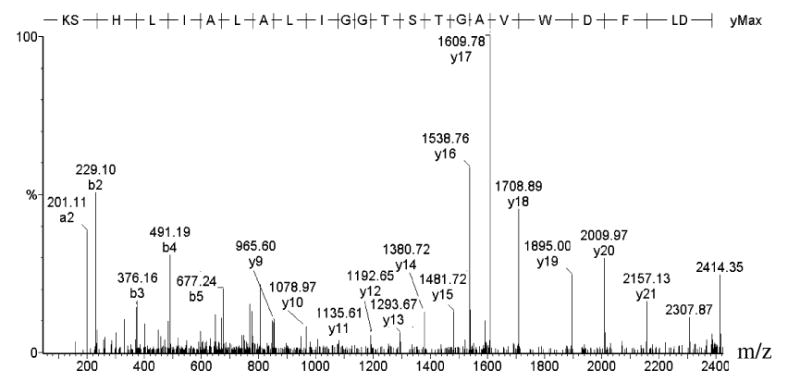
Tandem mass spectrum of iPLA2β tryptic peptide that contains the serine lipase consensus sequence (456DLFDWVA463GTSTG467GILALAILHSK478) produced by CAD of [M + 3H]3+ (m/z 795.41 Da). BEL-treated, His-tagged iPLA2β was digested with trypsin, and resultant peptides were analyzed as in Figure 2.
A formal possibility is that a modified peptide containing the lipase consensus sequence of iPLA2β is formed during BEL treatment but that the resultant peptide is not recovered from proteolytic digests or identified in LC/ESI/MS/MS analyses. To evaluate this possibility, we performed relative quantitative estimates of the abundance of peptide 456DLFDWVAGTSTGGILALAILHSK478 that contains the serine lipase consensus sequence site in untreated preparations of iPLA2β and in BEL-treated samples using the multiple normalization method of Steen et al. (53). In this method, the abundances of ions representing modified peptides are compared to those of ions that represent unmodified peptides. Here, the latter are those peptides not modified upon treating iPLA2β with BEL. Six tryptic peptides from iPLA2β that were routinely observed in proteolytic digests of BEL-treated and untreated iPLA2β, that were never observed in modified form, and that contained no cysteine residues were selected as the set of peptides that were not modified by BEL to which the abundance of modified peptides are compared (Table 1). This set of peptides represents regions of sequence from near the N-terminus, in the middle, and near the C-terminus of iPLA2β.
Table 1.
Relative Quantitation of Peptides 422SHDHLLCLDGGGVK435 and 456DLFDWVAGTSTGGILALAILHSK478 a
| control | BEL treated | ratio | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| start AA–end AA | m/z | Charge state | intensity | F422–435 | F456–478 | intensity | F422–435 | F456–478 | N422–435 | N456–478 | |
| set of reference internal peptides | |||||||||||
| 582SSGAAPTYFRPNGR595 | 740.8 | 2+ | 788 | 0.16 | 0.62 | 458 | 0.18 | 0.65 | 1.12 | 1.05 | |
| 538QPAELHLFR546 | 555.8 | 2+ | 410 | 0.31 | 1.20 | 268 | 0.31 | 1.12 | 1.00 | 0.93 | |
| 596FLDGGLLANNPTLDAMTEIHEYNQDMIR623 | 1064.39 | 3+ | 534 | 0.24 | 0.92 | 396 | 0.21 | 0.76 | 0.88 | 0.82 | |
| 38VREEGQLILLQNASNR53 | 613.99 | 3+ | 633 | 0.20 | 0.77 | 476 | 0.17 | 0.63 | 0.87 | 0.81 | |
| 396QLQDLMPVSR405 | 593.74 | 2+ | 1190 | 0.11 | 0.41 | 877 | 0.09 | 0.34 | 0.88 | 0.83 | |
| 7LVNTLSSVTNLFSNPFR23 | 954.96 | 2+ | 584 | 0.22 | 0.84 | 368 | 0.22 | 0.81 | 1.03 | 0.97 | |
| quantitated peptides | |||||||||||
| 422SHDHLLCLDGGGVK435 | 725.79 | 2+ | 126 | 82 | av (N): | 0.96 | 0.90 | ||||
| 456DLFDWVAGTSTGGILALAILHSK478 | 795.41 | 3+ | 490 | 299 | std EV (N): | 0.10 | 0.10 | ||||
F422–435 (intensity fraction of peptide 422–435 on the internal peptides) is calculated by dividing the intensity of peptide 422–435 by the intensities of internal peptides. F456–478 (intensity fraction of peptide 456–478 on the internal peptides) is calculated by dividing the intensity of peptide 456–478 by the intensities of internal peptides. N422–435 (normalized relative content of peptide 422–435) is calculated by F422–435(BEL)/F422–435(control). N456–478 (normalized relative content of peptide 456–478) is calculated by F456–478(BEL)/F456–478(control).
Table 1 summarizes raw and normalized intensity data for that set of reference peptides and for peptides to which they were compared in untreated and BEL-treated preparations of iPLA2β. One peptide that was compared to the set of unmodified reference peptides is 456DLFDWVAGTSTGGILALAILHSK478, which contains the 463GTSTG467 serine lipase consensus. The relative abundance of that peptide in tryptic digests of BEL-treated vs untreated iPLA2β preparations is estimated to be 0.90 ± 0.10, which indicates that this peptide is nearly equally abundant in untreated and BEL-treated iPLA2β samples. This supports the conclusion that BEL does not form a stable adduct with S465 or other residues in the GTSTG serine lipase consensus site as its mechanism of inactivation of iPLA2β.
Facility with Which Various Cysteine Residues in the iPLA2β Sequence Are Modified by BEL Treatment
Another peptide whose abundance was compared to that of the set of unmodified reference peptides was 422S–K435. That peptide contains Cys428 and, as discussed above, is unusual among cysteine-containing iPLA2β tryptic peptides because its cysteine residue is never observed to be modified in BEL-treated iPLA2β. The relative abundance of 422S–K435 between BEL-treated and untreated iPLA2β preparations is 0.96 (Table 1), which supports the observation that Cys428 is not modified by BEL treatment of iPLA2β under conditions where many other cysteine residues do form stable adducts.
Table 2 illustrates that other cysteine residues that were not observed to be modified at any tested concentration of BEL include Cys680, Cys681, Cys143, and Cys153. These residues participate in intramolecular disulfide bonds that protect them from modification by BEL. Cys428 has not been observed to participate in disulfide bonds but nonetheless has never been observed to be modified in BEL-treated iPLA2β. We have been unable to observe a tryptic peptide that contains Cys175 by MALDI/TOF/MS or LC/ESI/MS/MS, and this region of iPLA2β sequence is among the 20% that is not represented among peptides identified in proteolytic digests.
Table 2.
BEL-Modified Peptides Identified by MS and MS/MS Analyses
| MW | AA | μM | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| theoretical | detected | Δ mass | start | end | miss | AA sequence | 0 | 0.5 | 1 | 2 | 5 | 10 | 20 |
| 1175.59 | 1429.83 | 254.24 | 54 | 63 | 0 | (R)TWDCVLVSPR(N) | − | − | − | − | + | + | + |
| 915.39 | 1169.41 | 254.02 | 133 | 139 | 0 | (R)ECFHHSR(I)a | − | + | + | + | + | + | + |
| 2359.06 | 2613.21 | 254.15 | 140 | 161 | 0 | (R)IISCANSTENEEGCTPLHLACR(K) | − | − | + | + | + | + | + |
| 2607.21 | 163 | 185 | 0 | (K)GDSEILVELVQYCHAQMDVTDNK(G) | mis | mis | mis | mis | mis | mis | mis | ||
| 2579.28 | 2834.51 | 254.23 | 208 | 232 | 0 | (K)NASAGLNQVNNQGLTPLHLACQMGK(Q) | − | − | + | + | + | + | + |
| 901.53 | 1155.77 | 254.24 | 238 | 245 | 0 | (R)VLLLCNAR(C) | − | + | + | + | + | + | + |
| 2164.05 | 2418.4 | 254.35 | 246 | 265 | 0 | (R)CNIMGPGGFPIHTAMKFSQK(F) | − | − | − | − | − | + | + |
| 1863.84 | 2118.13 | 254.29 | 266 | 282 | 0 | (K)GCAEMIISMDSNQIHSK(D) | − | − | + | + | + | + | + |
| 2060.96 | 2314.96 | 254.00 | 307 | 327 | 0 | (R)GCDVDSTSASGNTALHVAVTR(N) | − | − | − | − | + | + | + |
| 2071.02 | 2324.97 | 253.95 | 328 | 346 | 1 | (R)NRFDCVMVLLTYGANAGAR(G) | − | − | − | + | + | + | + |
| 1450.71 | 1450.58 | −0.13 | 422 | 435 | 0 | (R)SHDHLLCLDGGGVK(G) | − | − | − | − | − | − | − |
| 2291.20 | 2545.35 | 254.15 | 559 | 578 | 0 | (R)CTPNINLKPPTQPADQLVWR(A) | − | − | − | − | − | + | + |
| 2469.26 | 2723.56 | 254.30 | 644 | 665 | 0 | (K)SPQVPVTCVDVFRPSNPWELAK(T) | − | + | + | + | + | + | + |
| 1308.52 | 1308.51 | 0.01 | 676 | 687 | 0 | (K)MVVDCCTDPDGR(A) | D | D | D | D | D | D | D |
| 1502.69 | 1756.82 | 254.13 | 694 | 705 | 0 | (R)AWCEMVGIQYFR(L) | − | − | − | − | − | − | + |
Indicates the peptide was identified by MALDI/TOF only. Mis indicates the peptide was not detected either by MALDI/TOF or by Q-TOF. D denotes an internally disufide-linked peptide. AA denotes amino acid. Miss denotes a missed cleavage site. MW denotes molecular weight. C denotes a non-BEL-modified cysteine.
Aside from the above exceptions, most other iPLA2β cysteine residues were observed to be modified by BEL treatment, although some are modified only at rather high BEL concentrations (Table 2). Cys135, Cys243, and Cys651 are modified at BEL concentrations as low as 0.5 μM, while Cys697 is not modified at a BEL concentration of 10 μM but is modified at 20 μM BEL. Cys247 and Cys560 are modified at 10 μM but not 5 μM BEL. Cys309 is modified at 5 μM but not at 2 μM BEL. Cys333 is modified at 2 μM but not at 1 μM BEL. Cys144, Cys229, and Cys268 are modified at 1 μM but not at 0.5 μM BEL. There is thus a wide range of sensitivities to modification by various concentrations of BEL among different iPLA2β cysteine residues, and the most sensitive is Cys651, 20% of which is modified at 0.5 μM BEL. The data in Table 2 must be evaluated with the caveat that the concentration of BEL required to yield detectable amounts of a modified peptide will depend on analytic sensitivity, which could differ among different peptides and perhaps between nonmodified and modified peptide pairs.
BEL Concentration Dependence of Cysteine Modification and of iPLA2β Inactivation
Figure 5 illustrates the BEL concentration dependence of iPLA2β catalytic inactivation and loss of free cysteine thiol groups as estimated by a spectrophotometric assay with DTNB reagent (Figure 5A) and by mass spectrometric measurements of the relative abundance of unmodified and modified cysteine-containing peptides from tryptic digests of untreated and BEL-treated iPLA2β (Figure 5B–D). The cysteine free thiol content measured by spectrophotometry declined more than did iPLA2β activity at low BEL concentrations but declined less than did iPLA2β activity at higher BEL concentrations (Figure 5A). This might indicate that modifying many cysteines does not affect catalytic activity, but when one or a small number of critical cysteine residues is modified, activity falls precipitously as a steeper function of increasing BEL concentration than does protein bulk thiol content.
FIGURE 5.
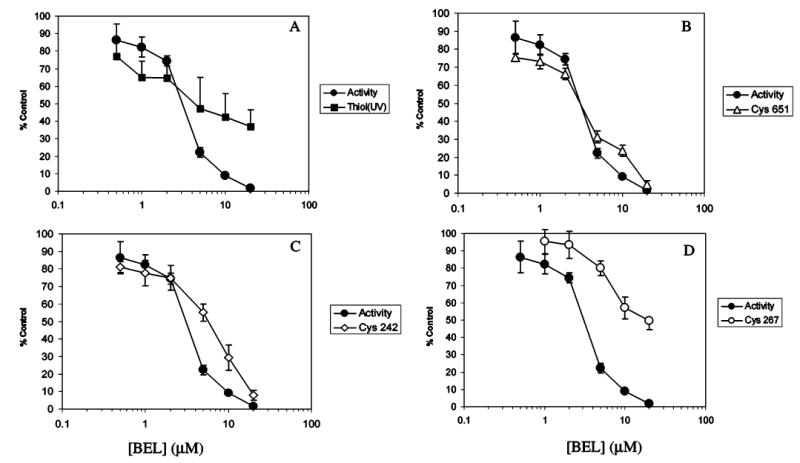
BEL concentration dependence of the decline in iPLA2β activity, free thiol content, and fraction of unmodified Cys651, Cys242, or Cys267. Purified, His-tagged iPLA2β was incubated (37 °C, 10 min) with various concentrations of BEL, and aliquots were removed for measurement of iPLA2β specific activity (closed circles, all panels), free thiol content determined by reaction with DTNB and spectrophotometry (closed squares, panel A), and fraction of unmodified Cys651 (open triangles, panel B), Cys242 (open diamonds, panel C), or Cys267 (open circles, panel D) determined by LC/MS/MS analyses of tryptic digests. Values are expressed as a percent of those observed at a BEL concentration of zero. Mean values (±SD) are displayed (n = 6).
There is a closer correspondence between decline in activity and decline in unmodified cysteine content estimated mass spectrometrically. Of all the iPLA2β cysteine residues that were observed to be modified, the BEL concentration dependence of the decline of unmodified Cys651 most closely follows the decline of iPLA2β activity (Figure 5B). Both curves in Figure 5B have an IC50 value of about 3.5 μM BEL, suggesting that modification of Cys651 might be involved in iPLA2β inactivation.
BEL Concentration Dependence of Inactivation of an iPLA2β Mutant Protein in Which Cys651 Is Replaced by Ala651
To evaluate further the possibility that alkylation of Cys651 might play a role in iPLA2β inactivation by BEL, that residue was replaced with Ala651, which has no thiol group that can be alkylated, by site-directed mutagenesis. The His-tagged mutant iPLA2βA651 protein was expressed in a baculovirus–Sf9 cell system and purified by immobilized metal affinity chromatography, in the manner illustrated for His-tagged native iPLA2β in Figure 1. The iPLA2β catalytic activity and the BEL concentration dependence of inactivation of the iPLA2βA651 mutant protein were then compared to native iPLA2β, as illustrated in Figure 6.
FIGURE 6.
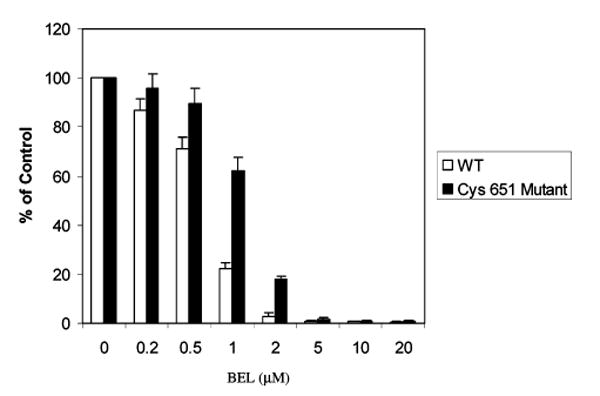
BEL concentration dependence of inactivation of an iPLA2β mutant protein in which Cys651 is replaced by Ala651. The Cys651 residue of iPLA2β was replaced with Ala651 by site-directed mutagenesis, and the recombinant iPLA2βA651 mutant protein was expressed in a baculovirus–Sf9 cell system and purified by immobilized metal affinity chromatography in the manner illustrated for native iPLA2β in Figure 1. The catalytic activity and sensitivity to inhibition by BEL of iPLA2βA651 (closed bars) and iPLA2β (open bars) were then determined as in Figures 1 and 5. Mean values (±SD) are displayed (n = 6).
The iPLA2βA651 mutant retained essentially full catalytic activity but was less sensitive to inhibition by BEL than was native iPLA2β (Figure 6). At a BEL concentration of 1 μM, for example, the iPLA2βA651 mutant protein exhibited about 60% of control activity, but native iPLA2β retained only about 20% of control activity. Although higher concentrations of BEL were required to inhibit iPLA2βA651 than to inhibit native iPLA2β, the mutant enzyme could be inactivated completely at sufficiently high BEL concentrations. These findings suggest that alkylation of Cys651 might be sufficient to inactivate iPLA2β but that Cys651 alkylation is not necessary iPLA2β inactivation, which can also result from modification of other residues.
Studies of the Mechanism of Covalent Modification of Cysteine Residues by BEL with Synthetic Cysteine-Containing Peptides
We had not anticipated our observation that numerous cysteine residues would be modified in BEL-treated iPLA2β because inactivation of chymotrypsin by BEL is reported to require covalent attachment of only a single BEL-derived moiety per enzyme molecule (45, 46). Moreover, BEL is thought to inflict its inactivating covalent modification on enzymes from a tethered position as an acyl intermediate adduct with the active site serine rather than by diffusing to reach distant targets (15, 16, 45, 46).
This would seem to constrain its site of action to a relatively small volume around the active site, which makes our observation that multiple cysteine residues throughout the iPLA2β sequence are modified by BEL seem puzzling. To test the possibility that a diffusible species derived from the action of iPLA2β on BEL mediates cysteine alkylation, two peptides (RGPCRAFL and PRCGVPDVA) that contain a free cysteine residue were synthesized and incubated with BEL in the presence or absence of iPLA2β.
Figure 7 illustrates that when these model peptides were incubated with BEL but not iPLA2β, only ions attributable to the unmodified peptides were observed in their MALDI/TOF mass spectra. The [M + H]+ ion of peptide RGPCRAFL is observed at the expected m/z 919.59 Da (Figure 7A) and that for peptide PRCGVPDVA at the expected m/z 913.67 Da (Figure 7B). This indicates that BEL cannot directly modify the peptides.
FIGURE 7.
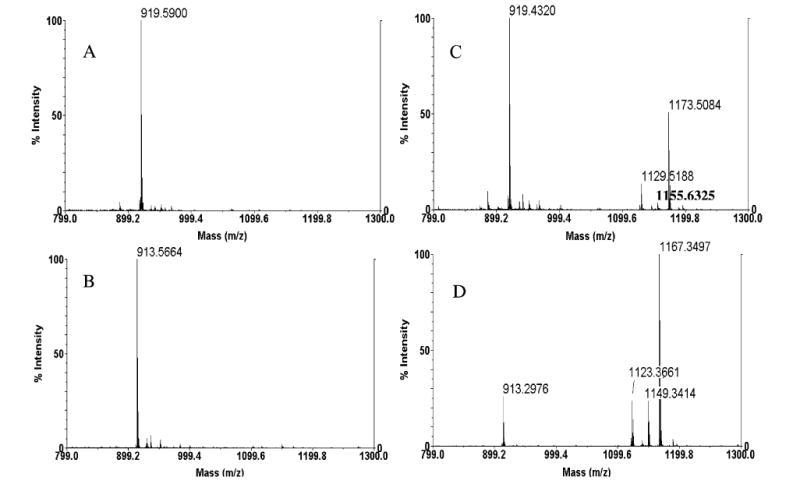
MALDI/TOF mass spectra of synthetic cysteine-containing peptides and their BEL-modified products. Synthetic peptides RGPCRAFL (panels A and C) and PRCGVPDVA (panels B and D) were incubated (30 min, 37 °C) with BEL (10 μM) in the absence (panels A and B) or presence (panels C and D) of iPLA2β (5 μg), and aliquots of the incubation mixtures were analyzed by MALDI/TOF/MS.
When the peptides were incubated with the same concentration of BEL in the presence of iPLA2β, adduct ions were observed for each peptide that reflected an increment in mass of 254 daltons, as expected for the previously observed cysteine adduct with a BEL-derived moiety (Figure 7C,D). The [M + 254 + H]+ ion for the modified peptide RGP(C-BEL)RAFL is observed at m/z 1173.66 (Figure 7C). Ions in this spectrum of m/z 1129.66 and 1155.63 reflect elimination from m/z 1173.66 of CO2 or H2O, respectively, from the BEL-derived moiety in the Cys adduct.
Similar ions were observed in the analogous spectrum for the peptide PR(C-BEL)GVPDVA derived from incubation of the parent peptide PRCGVPDVA with BEL and iPLA2β (Figure 7D). The [M + 254 + H]+ ion is observed at the expected m/z value of 1167.34, and losses of H2O or CO2, from that ion, yield the ions of m/z 1149.34 and 1123.37, respectively (Figure 7D). Figure 8 illustrates ESI/MS/MS spectra of BEL-modified peptides RGP(C-BEL)RAFL (m/z 1173.66) and PR(C-BEL)GVPDVA (m/z 1167.34), and the observed fragment ion series demonstrate that cysteine is the modified amino acid in peptides incubated with BEL and iPLA2β. The observations in Figures 7 and 8 demonstrate that catalytic action of iPLA2β on BEL is required to generate modified peptides from their parents.
FIGURE 8.
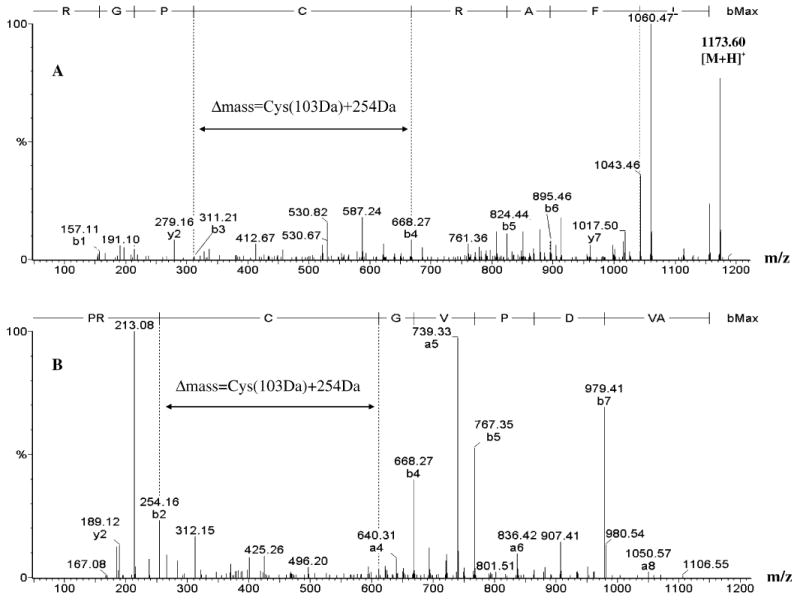
Tandem mass spectra of BEL-modified, synthetic, cysteine-containing peptides RGPCRAFL and PRCGVPDVA. Panel A is the ESI/MS/MS spectrum produced from CAD of ([M + 2H]2+ (m/z 586.80 Da) of peptide RGPCRAFL after incubation with iPLA2β and BEL, and panel B is the ESI/MS/MS spectrum produced from CAD of [M + 2H]2+ (m/z 583.71 Da) of peptide PRCGVPDVA after incubation with iPLA2β and BEL. The inset indicates that the mass difference between fragment ions generated before and after the modified cysteine is the residue mass of Cys (103 Da) plus 254 Da.
Figure 9 illustrates the BEL concentration dependence for modification of synthetic peptides in incubations with a fixed concentration of iPLA2β. The fraction of peptide molecules modified is estimated from the intensities of ions representing modified and unmodified forms of the peptides. This fraction increases with BEL concentrations up to 10 μM and declines at higher concentrations. This could reflect the opposing effects of production of BMKA from BEL to result in alkylation of bystander peptides but also in inactivation of iPLA2β. When all iPLA2β activity is eliminated, it can catalyze no further hydrolysis of BEL to the BMKA that alkylates bystander cysteine-containing peptides. Because iPLA2β is the source of BMKA, the local concentration of that species in the vicinity of iPLA2β could exceed that in the vicinity of bystander peptides, and this may result in its preferential alkylation at higher BEL concentrations.
FIGURE 9.
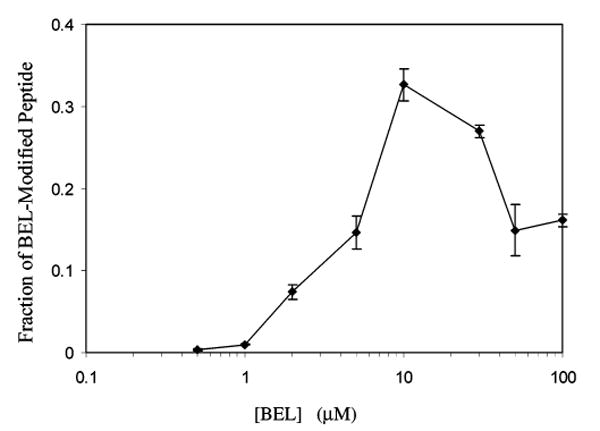
BEL concentration dependence of fractional modification of the synthetic, cysteine-containing peptide PR(C-BEL)GVPDVA. Peptide PRCGVPDVA was incubated (30 min, 37 °C) with various concentrations of BEL and 5 μg of iPLA2β, and the incubation mixture was analyzed by MALDI/TOF/MS. Fractional modification was calculated from intensities of spectral lines for modified and unmodified forms of the peptide. Mean values (±SD) are displayed (n = 6).
The observations in Figures 7–9 establish that alkylation of cysteine-containing peptides in incubations of BEL and iPLA2β requires the catalytic action of iPLA2β to generate a diffusible reactive species that can travel out of the active site to alkylate cysteine residues at distant sites, and this could also cause modification of cysteine residues in iPLA2β.
Protection of Cysteine Residues in iPLA2β from Modification upon Hydrolysis of BEL by Including Dithiothreitol in the Incubation Medium
To test the possibility that a diffusible alkylating species generated by iPLA2β-catalyzed BEL hydrolysis is responsible for modification of cysteine residues, the effect of including the scavenging nucleophile dithiothreitol (DTT) in the incubation mixtures was examined. Figure 10A illustrates that DTT suppresses alkylation of cysteine residues that otherwise occurs upon incubation of iPLA2β with BEL, and DTT also attenuates catalytic inactivation of iPLA2β. This supports the conclusion that modification of iPLA2β cysteine residues is mediated by a diffusible alkylating species that is accessible to and reacts with DTT in the incubation medium. Interestingly, most modifiable cysteine residues in iPLA2β were completely protected from BEL-induced modification at DTT concentrations between 0.2 and 1 mM. In contrast, Cys651 was protected from modification to a lesser extent at such DTT concentrations, and 5 mM DTT was required for complete protection. This residue thus could be less accessible to DTT-containing bulk solvent and buried within the folded enzyme, perhaps topologically close to the active site.
FIGURE 10.
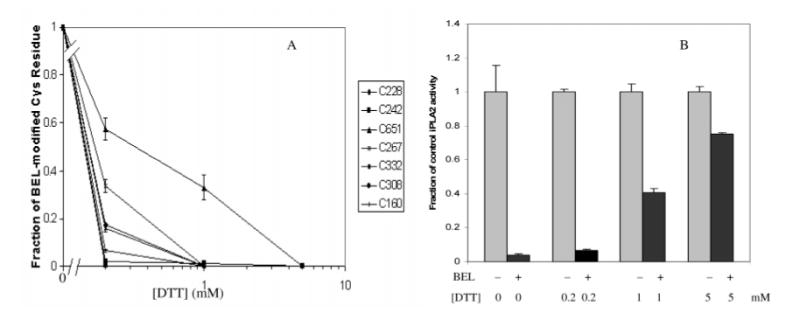
Effect of various concentrations of dithiothreitol (DTT) to protect iPLA2β from BEL-induced inactivation and alkylation of cysteine residues. Purified, His-tagged iPLA2β was incubated (37 °C, 10 min) without or with 4 μM BEL and various concentrations of DTT, and aliquots were removed for measurement of iPLA2β activity (panel A) or for tryptic digestion and LC/ESI/MS/MS analyses to identify peptides that contained modified and unmodified cysteine residues at various positions in the sequence (panel B). The fractional modification of various cysteine-containing peptides by BEL was determined by the multiple normalization method and expressed relative to the fraction in a sample without DTT (panel B). Mean values (±SD) are displayed (n = 6).
Discussion
Previous studies of covalent modification and catalytic inactivation of serine hydrolases by BEL and related haloenol lactones suggest that catalytic action of the hydrolase on BEL is required for inactivation and that covalent attachment of a BEL-derived moiety to the enzyme occurs (15, 16, 45, 46). Our results demonstrate that these features of the interaction of serine hydrolases and BEL pertain to recombinant iPLA2β and point to the unexpected conclusions that cysteine alkylation could be the inactivating covalent modification and could be mediated by a diffusible species.
The model of inhibition of serine hydrolases, e.g., chymotrypsin, is that BEL is a suicide substrate that forms an acyl-enzyme intermediate with the active site serine hydroxyl. This involves opening the lactone ring to yield a bromomethyl keto-substituted carboxylic acid ester. The bromomethyl keto moiety then alkylates a nearby nucleophile in a reaction that also generates HBr (45, 46). The acyl linkage between the BEL residue and the active site serine hydroxyl is then hydrolyzed to regenerate an unsubstituted active site serine and an alkyl keto acid residue of BEL that is covalently bound to the enzyme (45, 46). The initial acyl-enzyme structure formed between a serine hydrolase and an activated enol lactone inhibitor is stable in some cases, however, and the active site serine remains acylated (54). Our findings clearly indicate that this is not the case for iPLA2β because the active site S465 is not modified upon incubation with BEL under conditions where complete catalytic inactivation occurs.
Candidate nucleophiles that might react with the bromomethyl moiety derived from BEL include OH, SH, and NH groups in the side chains of several amino acids. Covalent linkages with haloenol latone suicide substrates have been reported to occur with serine and cysteine (47, 54) and perhaps with histidine (46) side chains. In the case of iPLA2β, cysteine is the only modified amino acid residue that we detect after incubation with BEL under conditions in which complete catalytic activation occurs and 80% overall sequence coverage is obtained in MS analyses of iPLA2β peptides in proteolytic digests.
In the 351T–P550 region of the linear amino acid sequence flanking the active site S465, combined data from trypsin and Glu-C digests of iPLA2β yield over 90% sequence coverage (Figure 11), and no modified amino acid residues in that region are observed in BEL-treated, catalytically inactivated iPLA2β. In addition, no modification of the cysteine residue closest to S465 in the linear amino acid sequence (Cys428) is observed in BEL-treated iPLA2β even under conditions where several other modified cysteine residues are observed. Proximity of amino acid residues in the linear amino acid sequence, of course, need not reflect their proximity in the topologic three-dimensional space of the correctly folded enzyme. Of the covalently modified cysteine residues that we identified, the BEL concentration dependence for Cys651 alkylation most closely parallels catalytic inactivation of iPLA2β. Although Cys651 is distant from the catalytic center S465 in the linear amino acid sequence of iPLA2β, it could be topologically proximate to that site in the correctly folded, catalytically active enzyme. This might cause it to be accessible to and readily alkylated by the bromomethyl keto moiety derived from BEL hydrolysis, and that alkylation could impair substrate binding or catalysis.
FIGURE 11.

Amino acid residues in the iPLA2β sequence identified in MALDI/TOF/MS and/or LC/ESI/MS/MS analyses of BEL-treated iPLA2β preparations digested with the endoproteases trypsin or Glu-C. Purified, His-tagged iPLA2β (10 μg) was treated (37 °C, 10 min) with BEL (20 μM), centrifugally filtered, and digested on-filter with trypsin or Glu-C. Digests were then analyzed by MALDI/TOF/MS and LC/ESI/MS/MS. Residues contained in peptides identified in LC/ESI/MS/MS analyses are denoted by italicized, bold type. Those identified in MALDI/TOF/MS analyses are denoted by bold, nonitalicized type. Residues that were not identified are denoted by plain type.
We believe that alkylation of cysteine residues involves the diffusible BEL hydrolysis product BMKA produced by iPLA2β catalysis, as illustrated in Scheme 1, rather than a bromomethyl.keto moiety tethered to the active site serine. Consistent with this scheme, the mass of the cysteine adduct in peptides modified in incubations of BEL and iPLA2β is 254 Da greater than the unmodified peptide mass. This reflects elimination of HBr (81 Da) from BEL (317 Da) and addition of H2O (18 Da) to generate the BMKA that alkylates cysteine. BMKA itself irreversibly inactivates chymotrypsin (46) and thus has sufficient intrinsic reactivity to modify nucleophilic sites in enzymes and to cause catalytic inactivation.
Scheme 1.
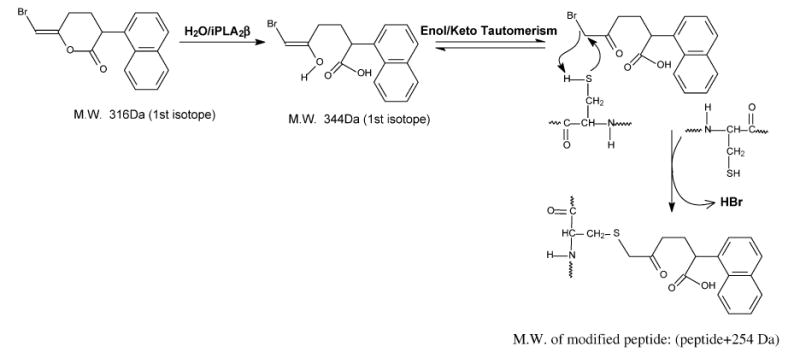
Scheme for Modification of Cysteine Residues by iPLA2β-Catalyzed BEL Hydrolysisa
The fact that the alkylating species generated from BEL by iPLA2β is diffusible rather than tethered to the active site serine is reflected by our findings that most iPLA2β cysteine residues can be alkylated upon BEL treatment, even though a much smaller fraction of them would be expected to be accessible to a bromomethyl keto moiety tethered to S465; that cysteine residues in bystander synthetic peptides are also alkylated when incubated with BEL and iPLA2β but not with BEL alone; and that adding DTT to the medium protects iPLA2β cysteine residues from alkylation during incubation with BEL.
That the BEL-derived alkylating species need not always diffuse from the region near the active site, enter the bulk solution, and travel to distant sites to inhibit iPLA2 is suggested by reports that under some conditions scavenging nucleophiles in the incubation medium do not prevent iPLA2 inhibition by BEL, that adding BMKA itself to the incubation does not inhibit iPLA2, and that a nearly 1:1 stoichiometry of covalently bound BEL moiety and inactivated iPLA2 molecules is observed (15, 16). Those studies, however, were performed before iPLA2β had been cloned and its sequence determined and before its molecular weight was known. Our studies here are the first of which we are aware to examine the mechanism of inhibition of purified, recombinant iPLA2β by BEL.
Alkylation of bystander peptides incubated with iPLA2β and BEL by the diffusible alkylating species BMKA suggests that other enzymes in the vicinity of iPLA2β exposed to BEL might be alkylated. Some alkylations could be functionally silent, as suggested by the poor correspondence between loss of bulk thiol groups and iPLA2β inactivation by BEL demonstrated in Figure 5, but other alkylations could inactivate bystander enzymes.
Our observation that cysteine is the amino acid most readily alkylated by BMKA suggests that enzymes with an active site cysteine residue might be among the most susceptible to inhibition by BEL or similar compounds. Glutathione S-transferase has a critical active site Cys47, and that enzyme is inactivated by a bromoenol lactone hydrolysis product by a mechanism that involves Cys47 alkylation (47). Similarly, the lipid phosphatase phosphatidate phosphohydrolase 1 (PAPH-1) is one non-iPLA2β enzyme recognized to be inhibited in BEL-treated cells, and that enzyme is also inhibited by other thiol-modifying agents (43).
Inhibition of PAPH-1 accounts for some biological effects of BEL (44), and active site cysteine residues are required for catalysis by many enzymes, including other lipid phosphatases (55), protein tyrosine phosphatases (56), and serine–threonine phosphatases (57) that are involved in insulin signaling, MAP kinase signaling, and tumor suppression (55–58). Our findings suggest that BEL should be used with appropriate caution when studying such events.
Acknowledgments
We thank Dr. Kevin Yarasheski for stimulating discussions about the relative quantitation method; Dr. Mary Wohltmann, Alan Bohrer, Wu Jin, and Sheng Zhang for excellent technical assistance; and Jessica Jackson for assistance with the manuscript.
Footnotes
This work was supported by U.S. Public Health Service Grants R37-DK34388, RO1-69455, P01-HL57278, P41-RR00954, P60-DK20579, and P30-DK56341.
Abbreviations: BEL, bromoenol lactone suicide substrate; BSA, bovine serum albumin; CAD, collisionally activated dissociation; ESI, electrospray ionization; HBSS, Hank's balanced salt solution; iPLA2β, group VIA phospholipase A2; kDa, kilodaltons; KRB, Krebs–Ringer bicarbonate buffer; MEM, minimal essential medium; MS, mass spectrometry; MS/MS, tandem mass spectrometry; Cap-LC, capillary high-performance liquid chromatography; PAGE, polyacrylamide gel electrophoresis; PAPH, phosphatidate phosphohydrolase; PLA2, phospholipase A2; RP-HPLC, reverse-phase high-performance liquid chromatography; RT, reverse transcriptase; SDS, sodium dodecyl sulfate; BMKA, bromomethyl keto acid.
BEL is hydrolyzed by iPLA2β to yield the bromomethyl keto acid BMKA. BMKA then alkylates the thiol group of cysteine residues by nucleophilic addition to eliminate HBr and form a thioether linkage to an α-keto-substituted carboxylic acid.
References
- 1.Brash AR. Arachidonic acid as a bioactive molecule. J Clin Invest. 2001;107:1339–1345. doi: 10.1172/JCI13210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Radu CG, Yang LV, Riedinger M, Au M, Witte ON. T cell chemotaxis to lysophosphatidylcholine through the G2A receptor. Proc Natl Acad Sci USA. 2004;101:245–250. doi: 10.1073/pnas.2536801100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Six D, Dennis E. The expanding superfamily of phospholipase A2 enzymes: classification and characterization. Biochim Biophys Acta. 2000;1488:1–19. doi: 10.1016/s1388-1981(00)00105-0. [DOI] [PubMed] [Google Scholar]
- 4.Ma Z, Turk J. The molecular biology of the group VIA Ca2+-independent phospholipase A2. Prog Nucleic Acid Res Mol Biol. 2001;67:1–33. doi: 10.1016/s0079-6603(01)67023-5. [DOI] [PubMed] [Google Scholar]
- 5.Murphy RC, Fitzpatrick FA. Arachidonate related lipid mediators. Methods Enzymol. 1990;187:1–628. [PubMed] [Google Scholar]
- 6.Gijon M, Spencer D, Kaiser A, Leslie C. Role of phosphorylation sites and the C2 domain in regulation of cytosolic phospholipase A2. J Cell Biol. 1999;145:1219–1232. doi: 10.1083/jcb.145.6.1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Underwood KW, Song C, Kriz RW, Chang XJ, Knopf JL, Lin LL. A novel calcium-independent phospholipase A2, cPLA2γ, that is prenylated and contains homology to cPLA2. J Biol Chem. 2002;273:21926–21932. doi: 10.1074/jbc.273.34.21926. [DOI] [PubMed] [Google Scholar]
- 8.Pinckard RT, Strifle BA, Kramer RM, Sharp JD. Molecular cloning of two new human paralogs of 85 kDa cytosolic phospholipase A2. J Biol Chem. 1999;274:8823–8831. doi: 10.1074/jbc.274.13.8823. [DOI] [PubMed] [Google Scholar]
- 9.Song C, Chang XJ, Bean KM, Proia MS, Knopf JL, Kriz RW. Molecular characterization of cytosolic phosholipase A2β. J Biol Chem. 1999;274:17063–17067. doi: 10.1074/jbc.274.24.17063. [DOI] [PubMed] [Google Scholar]
- 10.Ohto T, Uozumi N, Hirabayashi T, Shimuzu T. Identification of novel cytosolic phospholipase A2S, murine cPLA2δ, ∈ and ζ, which form a gene cluster with cPLA2β. J Biol Chem. 2005;280:24576–24583. doi: 10.1074/jbc.M413711200. [DOI] [PubMed] [Google Scholar]
- 11.Tang J, Kriz RW, Wolfman N, Shaffer M, Seehra J, Jones SS. A novel cytosolic calcium-independent phospholipase A2 contains eight ankyrin motifs. J Biol Chem. 1997;272:8567–8575. doi: 10.1074/jbc.272.13.8567. [DOI] [PubMed] [Google Scholar]
- 12.Balboa MA, Balsinde J, Jones S, Dennis EA. Identity between the calcium-independent phospholipase A2 enzymes from P388D1 macrophages and Chinese hamster ovary cells. J Biol Chem. 1997;272:8576–8590. doi: 10.1074/jbc.272.13.8576. [DOI] [PubMed] [Google Scholar]
- 13.Ma Z, Ramanadham S, Kempe K, Chi XS, Ladenson J, Turk J. Pancreatic islets express a Ca2+-independent phospholipase A2 enzyme that contains a repeated structural motif homologous to the integral membrane protein binding domain of ankyrin. J Biol Chem. 1997;272:11118–11127. [PubMed] [Google Scholar]
- 14.Balsinde J, Dennis EA. Function and inhibition of intracellular calcium-independent phospholipase A2. J Biol Chem. 1997;272:16069–16072. doi: 10.1074/jbc.272.26.16069. [DOI] [PubMed] [Google Scholar]
- 15.Hazen SL, Zupan LA, Weiss RH, Getman DP, Gross RW. Suicide inhibition of canine myocardial cytosolic calcium-independent phospholipase A2. Mechanism-based discrimination between calcium-dependent and -independent phospholipases A2. J Biol Chem. 1991;266:7227–7232. [PubMed] [Google Scholar]
- 16.Zupan LA, Weiss RH, Hazen S, Parnas BL, Aston KW, Lennon PJ, Getman DP, Gross RW. Structural determinants of haloenol lactone-mediated suicide inhibition of canine myocardial calcium-independent phospholipase A2. J Med Chem. 1993;36:95–100. doi: 10.1021/jm00053a012. [DOI] [PubMed] [Google Scholar]
- 17.Ma Z, Ramanadham S, Hu Z, Turk J. Cloning and expression of a type IV cytosolic phospholipase A2 enzyme from a pancreatic islet cDNA library. Comparison of the expressed activity with that of the islet type VI cytosolic phospholipase A2. Biochim Biophys Acta. 1998;1391:384–400. doi: 10.1016/s0005-2760(98)00027-7. [DOI] [PubMed] [Google Scholar]
- 18.Balsinde J, Dennis EA. Distinct roles in signal transduction for each of the phospholipase A2 enzymes present in P388D1 macrophages. J Biol Chem. 1996;271:6758–6765. doi: 10.1074/jbc.271.12.6758. [DOI] [PubMed] [Google Scholar]
- 19.Mancuso D, Jenkins CM, Gross RW. The genomic organization, complete mRNA sequence, cloning, and expression of a novel human intracellular membrane-associated calcium-independent phospholipase A2. J Biol Chem. 2000;275:9937–9945. doi: 10.1074/jbc.275.14.9937. [DOI] [PubMed] [Google Scholar]
- 20.Tanaka H, Takeya R, Sumimoto H. A novel intracellular membrane-bound calcium-independent phospholipase A2. Biochem Biophys Res Commun. 2000;272:320–326. doi: 10.1006/bbrc.2000.2776. [DOI] [PubMed] [Google Scholar]
- 21.Yang J, Han X, Gross RW. Identification of hepatic peroxisomal phospholipase A2 and characterization of arachidonic acid-containing choline glycerophospholipids in hepatic peroxisomes. FEBS Lett. 2003;546:247–250. doi: 10.1016/s0014-5793(03)00581-7. [DOI] [PubMed] [Google Scholar]
- 22.Mancuso DJ, Jenkins CM, Sims HF, Cohen JM, Yang J, Gross RW. Complex transcriptional and translational regulation of iPLA2γ resulting in multiple gene products containing dual competing sites for mitochondrial or peroxisomal localization. Eur J Biochem. 2004;271:4709–4724. doi: 10.1111/j.1432-1033.2004.04435.x. [DOI] [PubMed] [Google Scholar]
- 23.Van TM, Atkins J, Li Y, Glynn P. Human neuropathy target esterase catalyzes hydrolysis of membrane lipids. J Biol Chem. 2002;277:20942–209948. doi: 10.1074/jbc.M200330200. [DOI] [PubMed] [Google Scholar]
- 24.Jenkins CM, Mancuso DJ, Yan W, Sims HF, Gibson B, Gross RW. Identification, cloning, expression, and purifications of three novel human calcium-independent phospholipase A2 family members possessing triacylglycerol lipase and acylglycerol transacylase activities. J Biol Chem. 2004;279:48968–48975. doi: 10.1074/jbc.M407841200. [DOI] [PubMed] [Google Scholar]
- 25.Balsinde J, Bianco ID, Ackerman EJ, Conde-Friebos K, Dennis EA. Inhibition of calcium-independent phospholipase A2 prevents arachidonic acid incorporation and phospholipid remodeling in P388D1 macrophages. Proc Natl Acad Sci USA. 1995;92:8527–8531. doi: 10.1073/pnas.92.18.8527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Baburina I, Jackowski S. Cellular responses to excess phospholipid. J Biol Chem. 1999;274:9400–9408. doi: 10.1074/jbc.274.14.9400. [DOI] [PubMed] [Google Scholar]
- 27.Larsson PKA, Claesson HE, Kennedy BP. Multiple splice variants of the human calcium-independent phospholipase A2 and their effect on enzyme activity. J Biol Chem. 1998;273:207–214. doi: 10.1074/jbc.273.1.207. [DOI] [PubMed] [Google Scholar]
- 28.Ma Z, Wang X, Nowatzke W, Ramanadham S, Turk J. Human pancreatic islets express mRNA species encoding two distinct catalytically active isoforms of group VI phospholipase A2 (iPLA2) that arise from an exon-skipping mechanism of alternative splicing of the transcript from the iPLA2 gene on chromosome 22q13.1. J Biol Chem. 1999;274:9607–9616. doi: 10.1074/jbc.274.14.9607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Balboa MA, Saez Y, Balsinde J. Calcium-independent phospholipase A2 is required for lysozyme secretion in U937 promonocytes. J Immunol. 2003;170:5276–5280. doi: 10.4049/jimmunol.170.10.5276. [DOI] [PubMed] [Google Scholar]
- 30.Owada S, Larsson O, Arkhammar P, Katz AI, Chibalin AV, Berggren PO, Bertorello AM. Glucose decreases Na+,K+-ATPase activity in pancreatic beta-cells. An effect mediated via Ca2+-independent phospholipase A2 and protein kinase C-dependent phosphorylation of the alpha-subunit. J Biol Chem. 1999;274:2000–2008. doi: 10.1074/jbc.274.4.2000. [DOI] [PubMed] [Google Scholar]
- 31.Ramanadham S, Song H, Hsu FF, Zhang S, Crankshaw M, Grant G, Newgard C, Turk J. Pancreatic islets and insulinoma cells express a novel isoform of group VIA phospholipase A2 (iPLA2β) that participates in glucose-stimulated insulin secretion and is not produced by alternate splicing of the iPLA2β transcript. Biochemistry. 2003;42:13929–13940. doi: 10.1021/bi034843p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Atsumi GI, Murakami M, Kojima K, Hadano A, Tajima M, Kudo I. Distinct roles of two intracellular phospholipase A2s in fatty acid release in the cell death pathway. Proteolytic fragment of type IVA cytosolic phospholipase A2alpha inhibits stimulus-induced arachidonate release, whereas that of type VI Ca2+-independent phospholipase A2 augments spontaneous fatty acid release. J Biol Chem. 2000;275:18248–18258. doi: 10.1074/jbc.M000271200. [DOI] [PubMed] [Google Scholar]
- 33.Ramanadham S, Hsu FF, Zhang S, Jin C, Bohrer A, Ma Z, Turk J. Involvement of the group VIA phospholipase A2 (iPLA2β) in endoplasmic reticulum stress-induced apoptosis in insulinoma cells. Biochemistry. 2004;43:918–930. doi: 10.1021/bi035536m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jenkins CM, Han X, Mancuso DJ, Gross RW. Identification of calcium-independent phospholipase A2β (iPLA2β), and not iPLA2γ as the mediator of arginine vasopressin-induced arachidonic acid release in A-10 smooth muscle cells. Enantioselective mechanism-based discrimination of mammalian iPLA2s. J Biol Chem. 2002;277:32807–32814. doi: 10.1074/jbc.M202568200. [DOI] [PubMed] [Google Scholar]
- 35.Smani T, Zakharov SI, Csutora P, Leno E, Trepakova ES, Bolotina VM. A novel mechanism for the store-operated calcium influx pathway. Nat Cell Biol. 2004;6:113–120. doi: 10.1038/ncb1089. [DOI] [PubMed] [Google Scholar]
- 36.Williams SD, Ford DA. Calcium-independent phospholipase A2 mediates CREB phosphorylation and c-fos expression during ischemia. Am J Physiol. 2001;281:H168–H176. doi: 10.1152/ajpheart.2001.281.1.H168. [DOI] [PubMed] [Google Scholar]
- 37.Moran JM, Butler RM, McHowat J, Turk J, Wohltmann M, Gross RW, Corbett JA. Genetic and pharmacologic evidence that iPLA2β regulates virus-induced iNOS expression by macrophages. J Biol Chem. 2005;280:28162–28168. doi: 10.1074/jbc.M500013200. [DOI] [PubMed] [Google Scholar]
- 38.Murakami M, Kambe T, Shimbara S, Kudo I. Functional coupling between various phospholipase A2s and cyclooxygenases in immediate and delayed prostanoid biosynthetic pathways. J Biol Chem. 1999;274:3103–3115. doi: 10.1074/jbc.274.5.3103. [DOI] [PubMed] [Google Scholar]
- 39.Tay HR, Melendez AJ. FcγRI-triggered generation of arachidonic acid and eicosanoids requires iPLA2 but not cPLA2 in human monocytic cells. J Biol Chem. 2004;279:22505–22513. doi: 10.1074/jbc.M308788200. [DOI] [PubMed] [Google Scholar]
- 40.Ramanadham S, Ma Z, Arita H, Zhang S, Turk J. Type IB secretory phospholipase A2 is contained in the insulin secretory granules of pancreatic islet beta cells and is co-secreted with insulin upon stimulation of islets with glucose. Biochim Biophys Acta. 1998;1390:301–312. doi: 10.1016/s0005-2760(97)00189-6. [DOI] [PubMed] [Google Scholar]
- 41.Su X, Mancuso DJ, Bickel PE, Jenkins CM, Gross RW. Small interfering RNA knockdown of calcium-independent phospholipases A2β or γ inhibits the hormone-induced differentiation of 3T3-L1 preadipocytes. J Biol Chem. 2004;279:21740–21748. doi: 10.1074/jbc.M314166200. [DOI] [PubMed] [Google Scholar]
- 42.Shrai Y, Balsinde J, Dennis DA. Localization and functional interrelationships among cytosolic group IV, secreted group V, and Ca2+-independent group VI phospholipases A2s in P388D1 macrophages using GFP/RFP constructs. Biochim Biophys Acta. 2005;1735:119–129. doi: 10.1016/j.bbalip.2005.05.005. [DOI] [PubMed] [Google Scholar]
- 43.Balsinde J, Dennis EA. Bromoenol lactone inhibits magnesium-dependent phosphatidate phosphohydrolase and blocks triacylglycerol biosynthesis in mouse P388D1 macrophages. J Biol Chem. 1996;271:31937–31941. doi: 10.1074/jbc.271.50.31937. [DOI] [PubMed] [Google Scholar]
- 44.Fuentes L, Perez R, Nieto ML, Balsinde J, Balboa MA. Bromoenol lactone promotes cell death by a mechanism involving phosphatidate phosphohydrolase-1 rather than calcium-independent phospholipase A2. J Biol Chem. 2003;278:44683–44690. doi: 10.1074/jbc.M307209200. [DOI] [PubMed] [Google Scholar]
- 45.Daniels SB, Cooner E, Sofia MJ, Chakravarty PK, Katzenellenbogen JA. Haloenol lactones. Potent enzyme-activated irreversible inhibitors for α-chymotrypsin. J Biol Chem. 1983;258:15046–15053. [PubMed] [Google Scholar]
- 46.Daniels SB, Katzenellengbogen JA. Halo enol lactones: studies on the nechanism of inactivation of alphachymotrypsin. Biochemistry. 1986;25:1436–1444. doi: 10.1021/bi00354a037. [DOI] [PubMed] [Google Scholar]
- 47.Mitchell AE, Zheng J, Hammock BD, Bello ML, Jones AD. Structural and functional consequences of haloenol lactone inactivation of murine and human glutathione S-transferase. Biochemistry. 1998;37:6752–6759. doi: 10.1021/bi971846r. [DOI] [PubMed] [Google Scholar]
- 48.Nowatzke W, Ramanadham S, Hsu FF, Ma Z, Bohrer A, Turk J. Mass spectrometric evidence that agents that cause loss of Ca2+ from intracellular compartments induce hydrolysis of arachidonic acid from pancreatic islet membrane phospholipids by a mechanism that does not require a rise in cytosolic Ca2+ concentration. Endocrinology. 1998;139:4073–4085. doi: 10.1210/endo.139.10.6225. [DOI] [PubMed] [Google Scholar]
- 49.O'Reilly DR, Miller LK, Luckow VA. Baculovirus Expression Vector: A Laboratory Manual. W. H. Freeman and Co.; New York: 1992. [Google Scholar]
- 50.Bao S, Jin C, Zhang S, Turk J, Ma Z, Ramanadham S. Tracking iPLA2β movements in response to stimulation with insulin secretagogues in INS-1 Cells. Diabetes. 2004;53:S186–S189. doi: 10.2337/diabetes.53.2007.s186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Song H, Hecimovic S, Goate A, Hsu FF, Bao S, Vidavsky I, Ramanadham S, Turk J. Characterization of N-terminal processing of group VIA phospholipase A2 and of potential cleavage sites of amyloid precursor protein constructs by automated identification of signature peptides in LC/MS/MS analyses of proteolytic digests. J Am Soc Mass Spectrom. 2004;12:1780–1793. doi: 10.1016/j.jasms.2004.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gross RW, Ramanadham S, Kruszka KK, Han X, Turk J. Rat and human pancreatic islet cells contain a calcium ion independent phospholipase A2 activity selective for hydrolysis of arachidonate, which is stimulated by adenosine triphosphate and is specifically localized to islet β-cells. Biochemistry. 1993;32:327–336. doi: 10.1021/bi00052a041. [DOI] [PubMed] [Google Scholar]
- 53.Steen H, Jebanathirajah JA, Springer M, Kirschner MW. Stable isotope-free relative and absolute quantitation of protein phosphorylation stoichiometry by MS. Proc Natl Acad Sci U S A. 2005;102:3948–3953. doi: 10.1073/pnas.0409536102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Baek DJ, Reed PE, Daniels SB, Katzenellenbogen JA. Alternate substrate inhibitors of an alpha-chymotrypsin: enantioselective interaction of aryl-substituted enol lactones. Biochemistry. 1990;29:4305–4311. doi: 10.1021/bi00470a007. [DOI] [PubMed] [Google Scholar]
- 55.Maehama T, Dixon JE. The tumor suppressor, petn.mmac1, dephosphorylates the lipid second messenger, phosphatidylinositol 3,4,5-trisphosphate. J Biol Chem. 1998;273:13375–13378. doi: 10.1074/jbc.273.22.13375. [DOI] [PubMed] [Google Scholar]
- 56.Andersen JN, Mortensen OH, Peters GH, Drake PG, Iversen LF, Olsen OH, Jansen PG, Andersen HS, Tonks NK, Moller NP. Structural and evolutionary relationships among protein tyrosine phosphatases. Mol Cell Biol. 2001;21:7117–7136. doi: 10.1128/MCB.21.21.7117-7136.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Keyse SM. Protein phosphatases and the regulation of mitogen-activated protein kinase signaling. Curr Opin Cell Biol. 2000;12:186–192. doi: 10.1016/s0955-0674(99)00075-7. [DOI] [PubMed] [Google Scholar]
- 58.Goldstein BJ, Mahadev K, Wu X. Redox paradox. Insulin action is facilitated by insulin-stimulated reactive oxygen species with multiple potential signaling targets. Diabetes. 2005;54:311–321. doi: 10.2337/diabetes.54.2.311. [DOI] [PMC free article] [PubMed] [Google Scholar]


