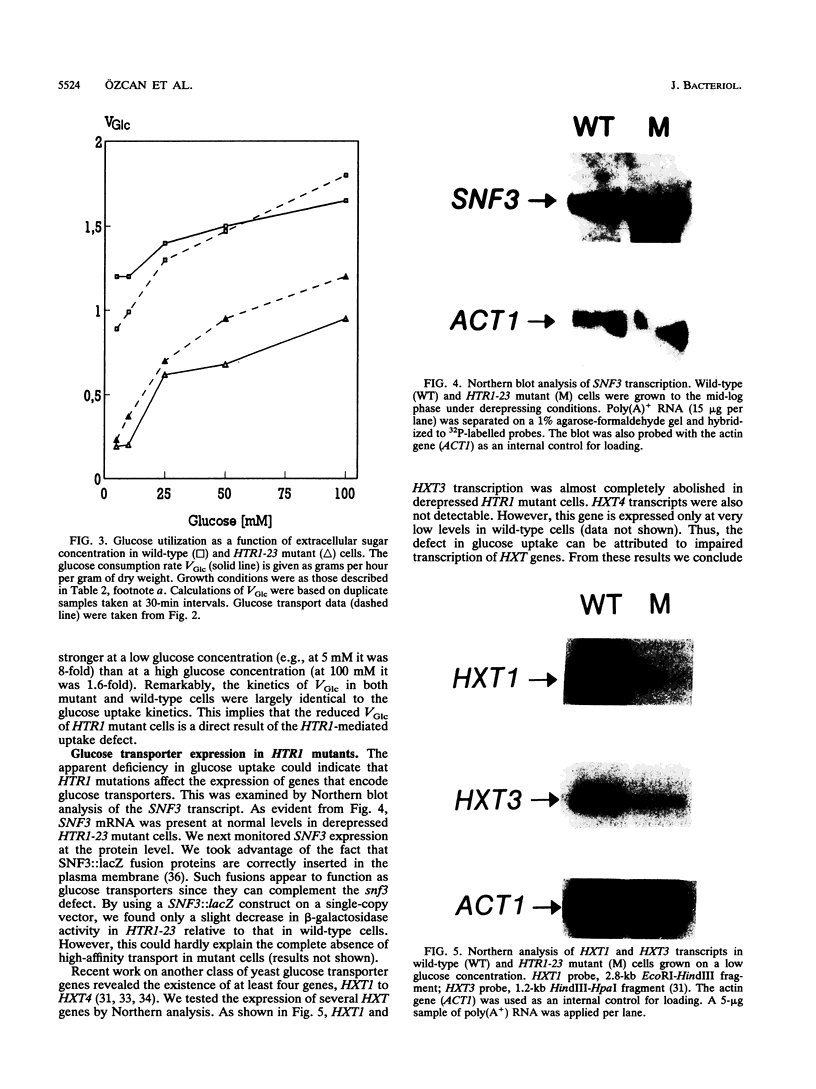
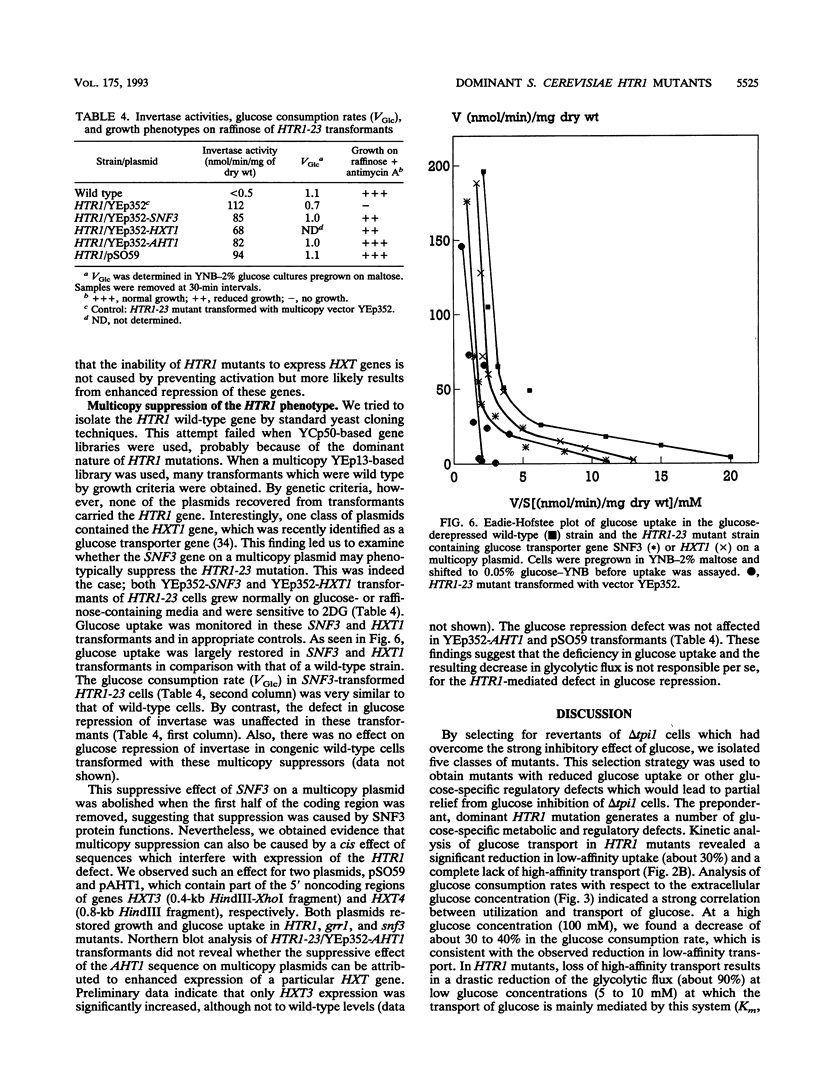
Abstract
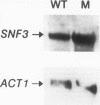
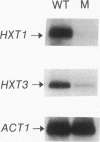
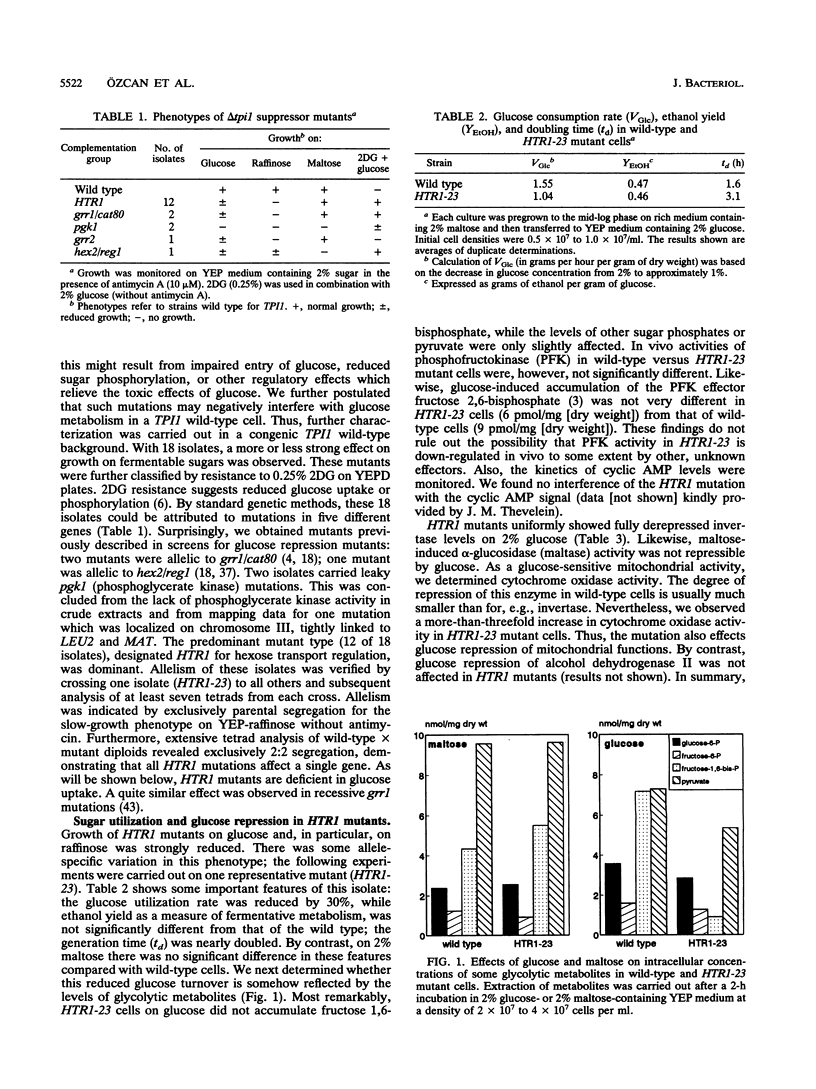
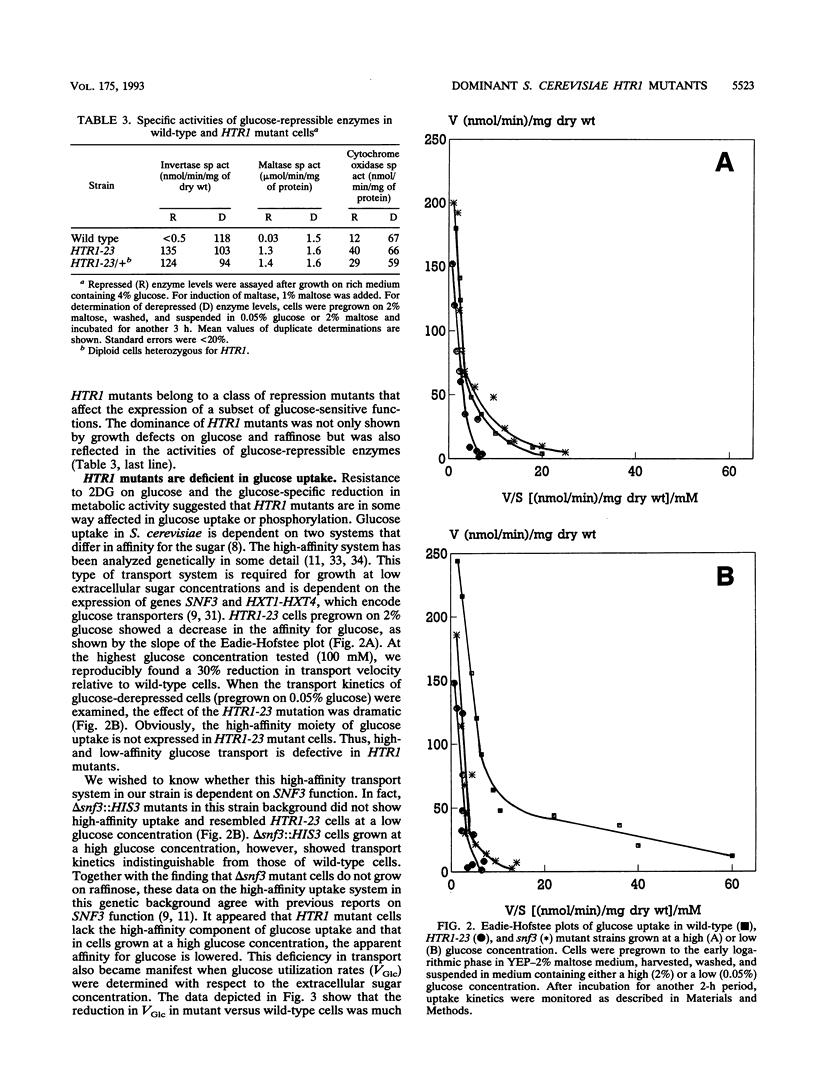
Growth and carbon metabolism in triosephosphate isomerase (delta tpi1) mutants of Saccharomyces cerevisiae are severely inhibited by glucose. By using this feature, we selected for secondary site revertants on glucose. We defined five complementation groups, some of which have previously been identified as glucose repression mutants. The predominant mutant type, HTR1 (hexose transport regulation), is dominant and causes various glucose-specific metabolic and regulatory defects in TPI1 wild-type cells. HTR1 mutants are deficient in high-affinity glucose uptake and have reduced low-affinity transport. Transcription of various known glucose transporter genes (HXT1, HXT3, and HXT4) was defective in HTR1 mutants, leading us to suggest that HTR mutations affect a negative factor of HXT gene expression. By contrast, transcript levels for SNF3, which encodes a component of high-affinity glucose uptake, were unaffected. We presume that HTR1 mutations affect a negative factor of HXT gene expression. Multicopy expression of HXT genes or parts of their regulatory sequences suppresses the metabolic defects of HTR1 mutants but not their derepressed phenotype at high glucose concentrations. This suggests that the glucose repression defect is not a direct result of the metabolically relevant defect in glucose transport. Alternatively, some unidentified regulatory components of the glucose transport system may be involved in the generation or transmission of signals for glucose repression.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Aguilera A. Mutations suppressing the effects of a deletion of the phosphoglucose isomerase gene PGI1 in Saccharomyces cerevisiae. Curr Genet. 1987;11(6-7):429–434. doi: 10.1007/BF00384603. [DOI] [PubMed] [Google Scholar]
- Alber T., Kawasaki G. Nucleotide sequence of the triose phosphate isomerase gene of Saccharomyces cerevisiae. J Mol Appl Genet. 1982;1(5):419–434. [PubMed] [Google Scholar]
- Avigad G. Stimulation of yeast phosphofructokinase activity by fructose 2,6-bisphosphate. Biochem Biophys Res Commun. 1981 Oct 15;102(3):985–991. doi: 10.1016/0006-291x(81)91635-1. [DOI] [PubMed] [Google Scholar]
- Bailey R. B., Woodword A. Isolation and characterization of a pleiotropic glucose repression resistant mutant of Saccharomyces cerevisiae. Mol Gen Genet. 1984;193(3):507–512. doi: 10.1007/BF00382091. [DOI] [PubMed] [Google Scholar]
- Biely P., Krátký Z., Bauer S. Metabolism of 2-deoxy-D glucose by Baker's yeast. IV. Incorporation of 2-deoxy-D-glucose into cell wall mannan. Biochim Biophys Acta. 1972 Feb 11;255(2):631–639. doi: 10.1016/0005-2736(72)90166-6. [DOI] [PubMed] [Google Scholar]
- Birnboim H. C., Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979 Nov 24;7(6):1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bisson L. F., Fraenkel D. G. Involvement of kinases in glucose and fructose uptake by Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1983 Mar;80(6):1730–1734. doi: 10.1073/pnas.80.6.1730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bisson L. F., Neigeborn L., Carlson M., Fraenkel D. G. The SNF3 gene is required for high-affinity glucose transport in Saccharomyces cerevisiae. J Bacteriol. 1987 Apr;169(4):1656–1662. doi: 10.1128/jb.169.4.1656-1662.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celenza J. L., Carlson M. A yeast gene that is essential for release from glucose repression encodes a protein kinase. Science. 1986 Sep 12;233(4769):1175–1180. doi: 10.1126/science.3526554. [DOI] [PubMed] [Google Scholar]
- Celenza J. L., Marshall-Carlson L., Carlson M. The yeast SNF3 gene encodes a glucose transporter homologous to the mammalian protein. Proc Natl Acad Sci U S A. 1988 Apr;85(7):2130–2134. doi: 10.1073/pnas.85.7.2130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciriacy M., Breitenbach I. Physiological effects of seven different blocks in glycolysis in Saccharomyces cerevisiae. J Bacteriol. 1979 Jul;139(1):152–160. doi: 10.1128/jb.139.1.152-160.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clifton D., Weinstock S. B., Fraenkel D. G. Glycolysis mutants in Saccharomyces cerevisiae. Genetics. 1978 Jan;88(1):1–11. doi: 10.1093/genetics/88.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dohmen R. J., Strasser A. W., Höner C. B., Hollenberg C. P. An efficient transformation procedure enabling long-term storage of competent cells of various yeast genera. Yeast. 1991 Oct;7(7):691–692. doi: 10.1002/yea.320070704. [DOI] [PubMed] [Google Scholar]
- Entian K. D., Barnett J. A. Regulation of sugar utilization by Saccharomyces cerevisiae. Trends Biochem Sci. 1992 Dec;17(12):506–510. doi: 10.1016/0968-0004(92)90341-6. [DOI] [PubMed] [Google Scholar]
- Entian K. D., Fröhlich K. U. Saccharomyces cerevisiae mutants provide evidence of hexokinase PII as a bifunctional enzyme with catalytic and regulatory domains for triggering carbon catabolite repression. J Bacteriol. 1984 Apr;158(1):29–35. doi: 10.1128/jb.158.1.29-35.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Entian K. D., Zimmermann F. K. Glycolytic enzymes and intermediates in carbon catabolite repression mutants of Saccharomyces cerevisiae. Mol Gen Genet. 1980 Jan;177(2):345–350. doi: 10.1007/BF00267449. [DOI] [PubMed] [Google Scholar]
- Entian K. D., Zimmermann F. K. New genes involved in carbon catabolite repression and derepression in the yeast Saccharomyces cerevisiae. J Bacteriol. 1982 Sep;151(3):1123–1128. doi: 10.1128/jb.151.3.1123-1128.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinberg A. P., Vogelstein B. "A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity". Addendum. Anal Biochem. 1984 Feb;137(1):266–267. doi: 10.1016/0003-2697(84)90381-6. [DOI] [PubMed] [Google Scholar]
- Flick J. S., Johnston M. GRR1 of Saccharomyces cerevisiae is required for glucose repression and encodes a protein with leucine-rich repeats. Mol Cell Biol. 1991 Oct;11(10):5101–5112. doi: 10.1128/mcb.11.10.5101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fröhlich K. U., Entian K. D., Mecke D. Cloning and restriction analysis of the hexokinase PII gene of the yeast Saccharomyces cerevisiae. Mol Gen Genet. 1984;194(1-2):144–148. doi: 10.1007/BF00383509. [DOI] [PubMed] [Google Scholar]
- Gancedo J. M. Carbon catabolite repression in yeast. Eur J Biochem. 1992 Jun 1;206(2):297–313. doi: 10.1111/j.1432-1033.1992.tb16928.x. [DOI] [PubMed] [Google Scholar]
- Goldstein A., Lampen J. O. Beta-D-fructofuranoside fructohydrolase from yeast. Methods Enzymol. 1975;42:504–511. doi: 10.1016/0076-6879(75)42159-0. [DOI] [PubMed] [Google Scholar]
- González M. I., Stucka R., Blázquez M. A., Feldmann H., Gancedo C. Molecular cloning of CIF1, a yeast gene necessary for growth on glucose. Yeast. 1992 Mar;8(3):183–192. doi: 10.1002/yea.320080304. [DOI] [PubMed] [Google Scholar]
- Gozalbo D. Multiple copies of SUC4 regulatory regions may cause partial de-repression of invertase synthesis in Saccharomyces cerevisiae. Curr Genet. 1992 May;21(6):437–442. doi: 10.1007/BF00351652. [DOI] [PubMed] [Google Scholar]
- Guarente L. Yeast promoters and lacZ fusions designed to study expression of cloned genes in yeast. Methods Enzymol. 1983;101:181–191. doi: 10.1016/0076-6879(83)01013-7. [DOI] [PubMed] [Google Scholar]
- Hill J. E., Myers A. M., Koerner T. J., Tzagoloff A. Yeast/E. coli shuttle vectors with multiple unique restriction sites. Yeast. 1986 Sep;2(3):163–167. doi: 10.1002/yea.320020304. [DOI] [PubMed] [Google Scholar]
- Ko C. H., Liang H., Gaber R. F. Roles of multiple glucose transporters in Saccharomyces cerevisiae. Mol Cell Biol. 1993 Jan;13(1):638–648. doi: 10.1128/mcb.13.1.638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruckeberg A. L., Bisson L. F. The HXT2 gene of Saccharomyces cerevisiae is required for high-affinity glucose transport. Mol Cell Biol. 1990 Nov;10(11):5903–5913. doi: 10.1128/mcb.10.11.5903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kötter P., Amore R., Hollenberg C. P., Ciriacy M. Isolation and characterization of the Pichia stipitis xylitol dehydrogenase gene, XYL2, and construction of a xylose-utilizing Saccharomyces cerevisiae transformant. Curr Genet. 1990 Dec;18(6):493–500. doi: 10.1007/BF00327019. [DOI] [PubMed] [Google Scholar]
- Lewis D. A., Bisson L. F. The HXT1 gene product of Saccharomyces cerevisiae is a new member of the family of hexose transporters. Mol Cell Biol. 1991 Jul;11(7):3804–3813. doi: 10.1128/mcb.11.7.3804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma H., Bloom L. M., Zhu Z. M., Walsh C. T., Botstein D. Isolation and characterization of mutations in the HXK2 gene of Saccharomyces cerevisiae. Mol Cell Biol. 1989 Dec;9(12):5630–5642. doi: 10.1128/mcb.9.12.5630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall-Carlson L., Celenza J. L., Laurent B. C., Carlson M. Mutational analysis of the SNF3 glucose transporter of Saccharomyces cerevisiae. Mol Cell Biol. 1990 Mar;10(3):1105–1115. doi: 10.1128/mcb.10.3.1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto K., Yoshimatsu T., Oshima Y. Recessive mutations conferring resistance to carbon catabolite repression of galactokinase synthesis in Saccharomyces cerevisiae. J Bacteriol. 1983 Mar;153(3):1405–1414. doi: 10.1128/jb.153.3.1405-1414.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers A. M., Tzagoloff A., Kinney D. M., Lusty C. J. Yeast shuttle and integrative vectors with multiple cloning sites suitable for construction of lacZ fusions. Gene. 1986;45(3):299–310. doi: 10.1016/0378-1119(86)90028-4. [DOI] [PubMed] [Google Scholar]
- Nasmyth K. A., Tatchell K. The structure of transposable yeast mating type loci. Cell. 1980 Mar;19(3):753–764. doi: 10.1016/s0092-8674(80)80051-1. [DOI] [PubMed] [Google Scholar]
- Nehlin J. O., Ronne H. Yeast MIG1 repressor is related to the mammalian early growth response and Wilms' tumour finger proteins. EMBO J. 1990 Sep;9(9):2891–2898. doi: 10.1002/j.1460-2075.1990.tb07479.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neigeborn L., Schwartzberg P., Reid R., Carlson M. Null mutations in the SNF3 gene of Saccharomyces cerevisiae cause a different phenotype than do previously isolated missense mutations. Mol Cell Biol. 1986 Nov;6(11):3569–3574. doi: 10.1128/mcb.6.11.3569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose M., Albig W., Entian K. D. Glucose repression in Saccharomyces cerevisiae is directly associated with hexose phosphorylation by hexokinases PI and PII. Eur J Biochem. 1991 Aug 1;199(3):511–518. doi: 10.1111/j.1432-1033.1991.tb16149.x. [DOI] [PubMed] [Google Scholar]
- Thevelein J. M. Fermentable sugars and intracellular acidification as specific activators of the RAS-adenylate cyclase signalling pathway in yeast: the relationship to nutrient-induced cell cycle control. Mol Microbiol. 1991 Jun;5(6):1301–1307. doi: 10.1111/j.1365-2958.1991.tb00776.x. [DOI] [PubMed] [Google Scholar]
- Tolbert N. E. Isolation of subcellular organelles of metabolism on isopycnic sucrose gradients. Methods Enzymol. 1974;31:734–746. doi: 10.1016/0076-6879(74)31077-4. [DOI] [PubMed] [Google Scholar]
- Trumbly R. J. Glucose repression in the yeast Saccharomyces cerevisiae. Mol Microbiol. 1992 Jan;6(1):15–21. doi: 10.1111/j.1365-2958.1992.tb00832.x. [DOI] [PubMed] [Google Scholar]
- Van Schaftingen E., Lederer B., Bartrons R., Hers H. G. A kinetic study of pyrophosphate: fructose-6-phosphate phosphotransferase from potato tubers. Application to a microassay of fructose 2,6-bisphosphate. Eur J Biochem. 1982 Dec;129(1):191–195. doi: 10.1111/j.1432-1033.1982.tb07039.x. [DOI] [PubMed] [Google Scholar]
- Walsh R. B., Clifton D., Horak J., Fraenkel D. G. Saccharomyces cerevisiae null mutants in glucose phosphorylation: metabolism and invertase expression. Genetics. 1991 Jul;128(3):521–527. doi: 10.1093/genetics/128.3.521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann F. K., Scheel I. Mutants of Saccharomyces cerevisiae resistant to carbon catabolite repression. Mol Gen Genet. 1977 Jul 7;154(1):75–82. doi: 10.1007/BF00265579. [DOI] [PubMed] [Google Scholar]