Abstract
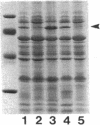
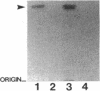
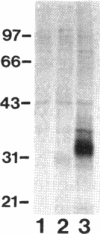
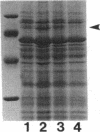
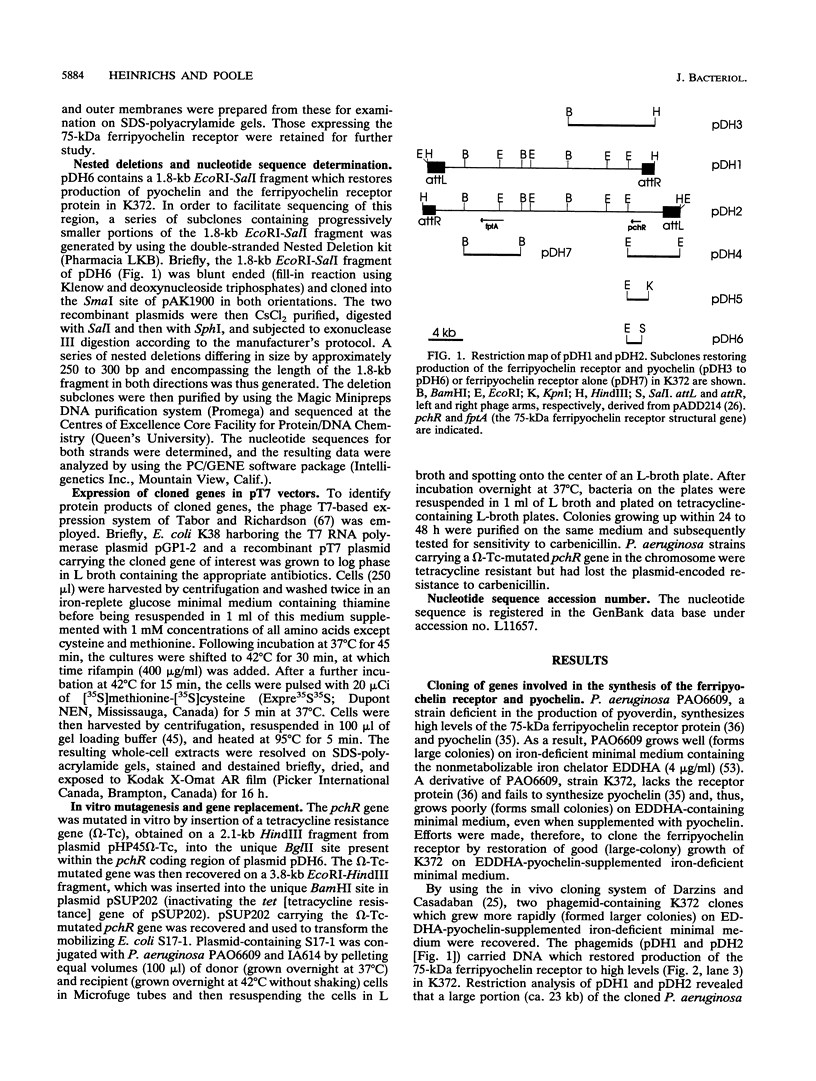
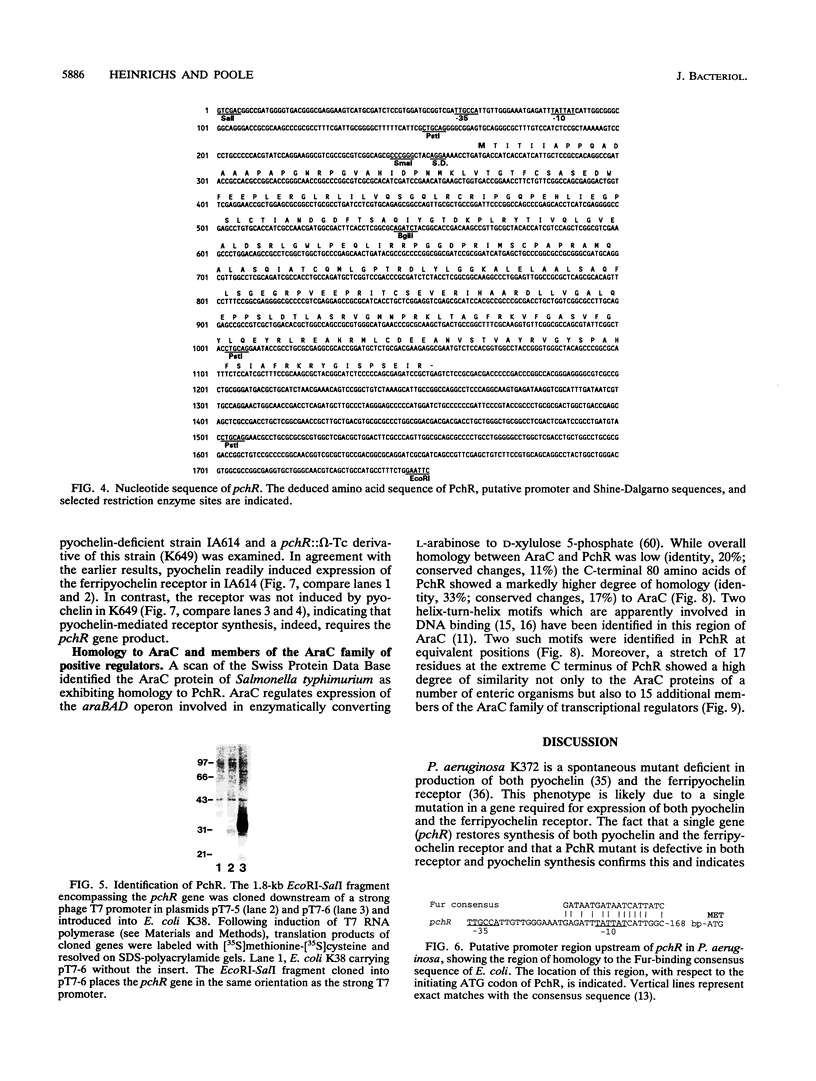
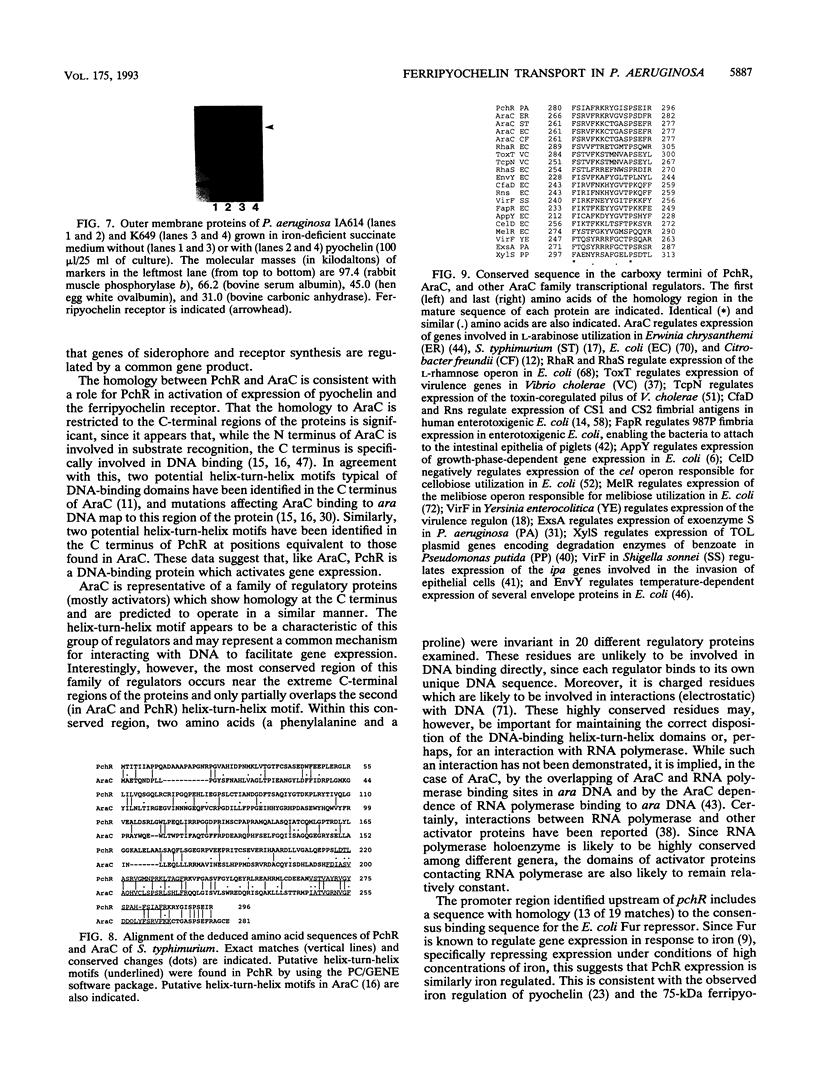
Pseudomonas aeruginosa K372 is deficient in the production of both the 75-kDa ferripyochelin receptor protein and pyochelin. A 1.8-kb EcoRI-SalI fragment which restored production of both the receptor protein and pyochelin was cloned. Nucleotide sequencing of the fragment revealed an open reading frame of 888 bp, designated pchR (pyochelin), capable of encoding a 296-amino-acid protein of a 32,339-Da molecular mass. By using a phage T7-based expression system, a protein of ca. 32 kDa was produced off the 1.8-kb fragment, confirming that this open reading frame was indeed expressed. A region exhibiting homology to the consensus Fur-binding site of Escherichia coli was identified upstream of the pchR coding region overlapping a putative promoter. In addition, the C-terminal 80 amino acid residues of PchR showed approximately 50% homology (identity, 31%; conserved changes, 19%) to the carboxy terminus of AraC, a known transcriptional activator of gene expression in E. coli, Salmonella typhimurium, Citrobacter freundii, and Erwinia chrysanthemi. Within the C-terminal region of PchR, AraC, and a number of other members of the AraC family of transcriptional activators, there exists a highly conserved 17-residue domain where, in fact, two residues are strictly maintained and two others exhibit only conserved changes, suggesting a common functional significance to this region in all of these proteins. These data are consistent with a role for PchR as a transcriptional activator of pyochelin and ferripyochelin receptor synthesis in P. aeruginosa. In agreement with this, a PchR mutant obtained by in vitro mutagenesis and gene replacement was deficient in production of the ferripyochelin receptor and pyochelin.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ankenbauer R. G. Cloning of the outer membrane high-affinity Fe(III)-pyochelin receptor of Pseudomonas aeruginosa. J Bacteriol. 1992 Jul;174(13):4401–4409. doi: 10.1128/jb.174.13.4401-4409.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ankenbauer R. G., Cox C. D. Isolation and characterization of Pseudomonas aeruginosa mutants requiring salicylic acid for pyochelin biosynthesis. J Bacteriol. 1988 Nov;170(11):5364–5367. doi: 10.1128/jb.170.11.5364-5367.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ankenbauer R., Hanne L. F., Cox C. D. Mapping of mutations in Pseudomonas aeruginosa defective in pyoverdin production. J Bacteriol. 1986 Jul;167(1):7–11. doi: 10.1128/jb.167.1.7-11.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ankenbauer R., Sriyosachati S., Cox C. D. Effects of siderophores on the growth of Pseudomonas aeruginosa in human serum and transferrin. Infect Immun. 1985 Jul;49(1):132–140. doi: 10.1128/iai.49.1.132-140.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atlung T., Nielsen A., Hansen F. G. Isolation, characterization, and nucleotide sequence of appY, a regulatory gene for growth-phase-dependent gene expression in Escherichia coli. J Bacteriol. 1989 Mar;171(3):1683–1691. doi: 10.1128/jb.171.3.1683-1691.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berry D., Kropinski A. M. Effect of lipopolysaccharide mutations and temperature on plasmid transformation efficiency in Pseudomonas aeruginosa. Can J Microbiol. 1986 May;32(5):436–438. doi: 10.1139/m86-082. [DOI] [PubMed] [Google Scholar]
- Botzenhart K., Rüden H. Hospital infections caused by Pseudomonas aeruginosa. Antibiot Chemother (1971) 1987;39:1–15. doi: 10.1159/000414328. [DOI] [PubMed] [Google Scholar]
- Brumlik M. J., Storey D. G. Zinc and iron regulate translation of the gene encoding Pseudomonas aeruginosa elastase. Mol Microbiol. 1992 Feb;6(3):337–344. doi: 10.1111/j.1365-2958.1992.tb01476.x. [DOI] [PubMed] [Google Scholar]
- Brunelle A., Schleif R. Determining residue-base interactions between AraC protein and araI DNA. J Mol Biol. 1989 Oct 20;209(4):607–622. doi: 10.1016/0022-2836(89)90598-6. [DOI] [PubMed] [Google Scholar]
- Burke K. A., Wilcox G. The araC gene of Citrobacter freundii. Gene. 1987;61(3):243–252. doi: 10.1016/0378-1119(87)90188-0. [DOI] [PubMed] [Google Scholar]
- Calderwood S. B., Mekalanos J. J. Iron regulation of Shiga-like toxin expression in Escherichia coli is mediated by the fur locus. J Bacteriol. 1987 Oct;169(10):4759–4764. doi: 10.1128/jb.169.10.4759-4764.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caron J., Coffield L. M., Scott J. R. A plasmid-encoded regulatory gene, rns, required for expression of the CS1 and CS2 adhesins of enterotoxigenic Escherichia coli. Proc Natl Acad Sci U S A. 1989 Feb;86(3):963–967. doi: 10.1073/pnas.86.3.963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cass L. G., Wilcox G. Mutations in the araC regulatory gene of Escherichia coli B/r that affect repressor and activator functions of AraC protein. J Bacteriol. 1986 Jun;166(3):892–900. doi: 10.1128/jb.166.3.892-900.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke P., Lee J. H., Burke K., Wilcox G. Mutations in the araC gene of Salmonella typhimurium LT2 which affect both activator and auto-regulatory functions of the AraC protein. Gene. 1992 Aug 1;117(1):31–37. doi: 10.1016/0378-1119(92)90486-9. [DOI] [PubMed] [Google Scholar]
- Clarke P., Lin H. C., Wilcox G. The nucleotide sequence of the araC regulatory gene in Salmonella typhimurium LT2. Gene. 1982 May;18(2):157–163. doi: 10.1016/0378-1119(82)90113-5. [DOI] [PubMed] [Google Scholar]
- Cornelis G., Sluiters C., de Rouvroit C. L., Michiels T. Homology between virF, the transcriptional activator of the Yersinia virulence regulon, and AraC, the Escherichia coli arabinose operon regulator. J Bacteriol. 1989 Jan;171(1):254–262. doi: 10.1128/jb.171.1.254-262.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox C. D., Adams P. Siderophore activity of pyoverdin for Pseudomonas aeruginosa. Infect Immun. 1985 Apr;48(1):130–138. doi: 10.1128/iai.48.1.130-138.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox C. D. Effect of pyochelin on the virulence of Pseudomonas aeruginosa. Infect Immun. 1982 Apr;36(1):17–23. doi: 10.1128/iai.36.1.17-23.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox C. D., Graham R. Isolation of an iron-binding compound from Pseudomonas aeruginosa. J Bacteriol. 1979 Jan;137(1):357–364. doi: 10.1128/jb.137.1.357-364.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox C. D. Iron reductases from Pseudomonas aeruginosa. J Bacteriol. 1980 Jan;141(1):199–204. doi: 10.1128/jb.141.1.199-204.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox C. D. Iron uptake with ferripyochelin and ferric citrate by Pseudomonas aeruginosa. J Bacteriol. 1980 May;142(2):581–587. doi: 10.1128/jb.142.2.581-587.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crosa J. H. The relationship of plasmid-mediated iron transport and bacterial virulence. Annu Rev Microbiol. 1984;38:69–89. doi: 10.1146/annurev.mi.38.100184.000441. [DOI] [PubMed] [Google Scholar]
- Darzins A., Casadaban M. J. In vivo cloning of Pseudomonas aeruginosa genes with mini-D3112 transposable bacteriophage. J Bacteriol. 1989 Jul;171(7):3917–3925. doi: 10.1128/jb.171.7.3917-3925.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darzins A., Casadaban M. J. Mini-D3112 bacteriophage transposable elements for genetic analysis of Pseudomonas aeruginosa. J Bacteriol. 1989 Jul;171(7):3909–3916. doi: 10.1128/jb.171.7.3909-3916.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean C. R., Poole K. Cloning and characterization of the ferric enterobactin receptor gene (pfeA) of Pseudomonas aeruginosa. J Bacteriol. 1993 Jan;175(2):317–324. doi: 10.1128/jb.175.2.317-324.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denoya C. D., Bechhofer D. H., Dubnau D. Translational autoregulation of ermC 23S rRNA methyltransferase expression in Bacillus subtilis. J Bacteriol. 1986 Dec;168(3):1133–1141. doi: 10.1128/jb.168.3.1133-1141.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fellay R., Frey J., Krisch H. Interposon mutagenesis of soil and water bacteria: a family of DNA fragments designed for in vitro insertional mutagenesis of gram-negative bacteria. Gene. 1987;52(2-3):147–154. doi: 10.1016/0378-1119(87)90041-2. [DOI] [PubMed] [Google Scholar]
- Francklyn C. S., Lee N. AraC proteins with altered DNA sequence specificity which activate a mutant promoter in Escherichia coli. J Biol Chem. 1988 Mar 25;263(9):4400–4407. [PubMed] [Google Scholar]
- Frank D. W., Iglewski B. H. Cloning and sequence analysis of a trans-regulatory locus required for exoenzyme S synthesis in Pseudomonas aeruginosa. J Bacteriol. 1991 Oct;173(20):6460–6468. doi: 10.1128/jb.173.20.6460-6468.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gensberg K., Hughes K., Smith A. W. Siderophore-specific induction of iron uptake in Pseudomonas aeruginosa. J Gen Microbiol. 1992 Nov;138(11):2381–2387. doi: 10.1099/00221287-138-11-2381. [DOI] [PubMed] [Google Scholar]
- Heinrichs D. E., Young L., Poole K. Pyochelin-mediated iron transport in Pseudomonas aeruginosa: involvement of a high-molecular-mass outer membrane protein. Infect Immun. 1991 Oct;59(10):3680–3684. doi: 10.1128/iai.59.10.3680-3684.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins D. E., Nazareno E., DiRita V. J. The virulence gene activator ToxT from Vibrio cholerae is a member of the AraC family of transcriptional activators. J Bacteriol. 1992 Nov;174(21):6974–6980. doi: 10.1128/jb.174.21.6974-6980.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochschild A., Irwin N., Ptashne M. Repressor structure and the mechanism of positive control. Cell. 1983 Feb;32(2):319–325. doi: 10.1016/0092-8674(83)90451-8. [DOI] [PubMed] [Google Scholar]
- Inouye S., Nakazawa A., Nakazawa T. Nucleotide sequence of the regulatory gene xylS on the Pseudomonas putida TOL plasmid and identification of the protein product. Gene. 1986;44(2-3):235–242. doi: 10.1016/0378-1119(86)90187-3. [DOI] [PubMed] [Google Scholar]
- Kato J., Ito K., Nakamura A., Watanabe H. Cloning of regions required for contact hemolysis and entry into LLC-MK2 cells from Shigella sonnei form I plasmid: virF is a positive regulator gene for these phenotypes. Infect Immun. 1989 May;57(5):1391–1398. doi: 10.1128/iai.57.5.1391-1398.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klaasen P., de Graaf F. K. Characterization of FapR, a positive regulator of expression of the 987P operon in enterotoxigenic Escherichia coli. Mol Microbiol. 1990 Oct;4(10):1779–1783. doi: 10.1111/j.1365-2958.1990.tb00556.x. [DOI] [PubMed] [Google Scholar]
- Lee N. L., Gielow W. O., Wallace R. G. Mechanism of araC autoregulation and the domains of two overlapping promoters, Pc and PBAD, in the L-arabinose regulatory region of Escherichia coli. Proc Natl Acad Sci U S A. 1981 Feb;78(2):752–756. doi: 10.1073/pnas.78.2.752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei S. P., Lin H. C., Heffernan L., Wilcox G. araB Gene and nucleotide sequence of the araC gene of Erwinia carotovora. J Bacteriol. 1985 Nov;164(2):717–722. doi: 10.1128/jb.164.2.717-722.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lugtenberg B., Meijers J., Peters R., van der Hoek P., van Alphen L. Electrophoretic resolution of the "major outer membrane protein" of Escherichia coli K12 into four bands. FEBS Lett. 1975 Oct 15;58(1):254–258. doi: 10.1016/0014-5793(75)80272-9. [DOI] [PubMed] [Google Scholar]
- Lundrigan M. D., Friedrich M. J., Kadner R. J. Nucleotide sequence of the Escherichia coli porin thermoregulatory gene envY. Nucleic Acids Res. 1989 Jan 25;17(2):800–800. doi: 10.1093/nar/17.2.800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menon K. P., Lee N. L. Activation of ara operons by a truncated AraC protein does not require inducer. Proc Natl Acad Sci U S A. 1990 May;87(10):3708–3712. doi: 10.1073/pnas.87.10.3708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neilands J. B. Microbial envelope proteins related to iron. Annu Rev Microbiol. 1982;36:285–309. doi: 10.1146/annurev.mi.36.100182.001441. [DOI] [PubMed] [Google Scholar]
- Neilands J. B. Microbial iron compounds. Annu Rev Biochem. 1981;50:715–731. doi: 10.1146/annurev.bi.50.070181.003435. [DOI] [PubMed] [Google Scholar]
- Ogierman M. A., Manning P. A. Homology of TcpN, a putative regulatory protein of Vibrio cholerae, to the AraC family of transcriptional activators. Gene. 1992 Jul 1;116(1):93–97. doi: 10.1016/0378-1119(92)90634-2. [DOI] [PubMed] [Google Scholar]
- Parker L. L., Hall B. G. Characterization and nucleotide sequence of the cryptic cel operon of Escherichia coli K12. Genetics. 1990 Mar;124(3):455–471. doi: 10.1093/genetics/124.3.455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poole K., Young L., Neshat S. Enterobactin-mediated iron transport in Pseudomonas aeruginosa. J Bacteriol. 1990 Dec;172(12):6991–6996. doi: 10.1128/jb.172.12.6991-6996.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prince R. W., Storey D. G., Vasil A. I., Vasil M. L. Regulation of toxA and regA by the Escherichia coli fur gene and identification of a Fur homologue in Pseudomonas aeruginosa PA103 and PA01. Mol Microbiol. 1991 Nov;5(11):2823–2831. doi: 10.1111/j.1365-2958.1991.tb01991.x. [DOI] [PubMed] [Google Scholar]
- Rogers H. J. Iron-Binding Catechols and Virulence in Escherichia coli. Infect Immun. 1973 Mar;7(3):445–456. doi: 10.1128/iai.7.3.445-456.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russel M., Model P. Replacement of the fip gene of Escherichia coli by an inactive gene cloned on a plasmid. J Bacteriol. 1984 Sep;159(3):1034–1039. doi: 10.1128/jb.159.3.1034-1039.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savelkoul P. H., Willshaw G. A., McConnell M. M., Smith H. R., Hamers A. M., van der Zeijst B. A., Gaastra W. Expression of CFA/I fimbriae is positively regulated. Microb Pathog. 1990 Feb;8(2):91–99. doi: 10.1016/0882-4010(90)90073-y. [DOI] [PubMed] [Google Scholar]
- Schnaitman C. A. Outer membrane proteins of Escherichia coli. 3. Evidence that the major protein of Escherichia coli O111 outer membrane consists of four distinct polypeptide species. J Bacteriol. 1974 May;118(2):442–453. doi: 10.1128/jb.118.2.442-453.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweizer H. P. Escherichia-Pseudomonas shuttle vectors derived from pUC18/19. Gene. 1991 Jan 2;97(1):109–121. doi: 10.1016/0378-1119(91)90016-5. [DOI] [PubMed] [Google Scholar]
- Sokol P. A. Surface expression of ferripyochelin-binding protein is required for virulence of Pseudomonas aeruginosa. Infect Immun. 1987 Sep;55(9):2021–2025. doi: 10.1128/iai.55.9.2021-2025.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokol P. A., Woods D. E. Demonstration of an iron-siderophore-binding protein in the outer membrane of Pseudomonas aeruginosa. Infect Immun. 1983 May;40(2):665–669. doi: 10.1128/iai.40.2.665-669.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sriyosachati S., Cox C. D. Siderophore-mediated iron acquisition from transferrin by Pseudomonas aeruginosa. Infect Immun. 1986 Jun;52(3):885–891. doi: 10.1128/iai.52.3.885-891.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabor S., Richardson C. C. A bacteriophage T7 RNA polymerase/promoter system for controlled exclusive expression of specific genes. Proc Natl Acad Sci U S A. 1985 Feb;82(4):1074–1078. doi: 10.1073/pnas.82.4.1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tobin J. F., Schleif R. F. Positive regulation of the Escherichia coli L-rhamnose operon is mediated by the products of tandemly repeated regulatory genes. J Mol Biol. 1987 Aug 20;196(4):789–799. doi: 10.1016/0022-2836(87)90405-0. [DOI] [PubMed] [Google Scholar]
- Vieira J., Messing J. The pUC plasmids, an M13mp7-derived system for insertion mutagenesis and sequencing with synthetic universal primers. Gene. 1982 Oct;19(3):259–268. doi: 10.1016/0378-1119(82)90015-4. [DOI] [PubMed] [Google Scholar]
- Wallace R. G., Lee N., Fowler A. V. The araC gene of Escherichia coli: transcriptional and translational start-points and complete nucleotide sequence. Gene. 1980 Dec;12(3-4):179–190. doi: 10.1016/0378-1119(80)90100-6. [DOI] [PubMed] [Google Scholar]
- Weber I. T., Steitz T. A. Model of specific complex between catabolite gene activator protein and B-DNA suggested by electrostatic complementarity. Proc Natl Acad Sci U S A. 1984 Jul;81(13):3973–3977. doi: 10.1073/pnas.81.13.3973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webster C., Kempsell K., Booth I., Busby S. Organisation of the regulatory region of the Escherichia coli melibiose operon. Gene. 1987;59(2-3):253–263. doi: 10.1016/0378-1119(87)90333-7. [DOI] [PubMed] [Google Scholar]