Abstract
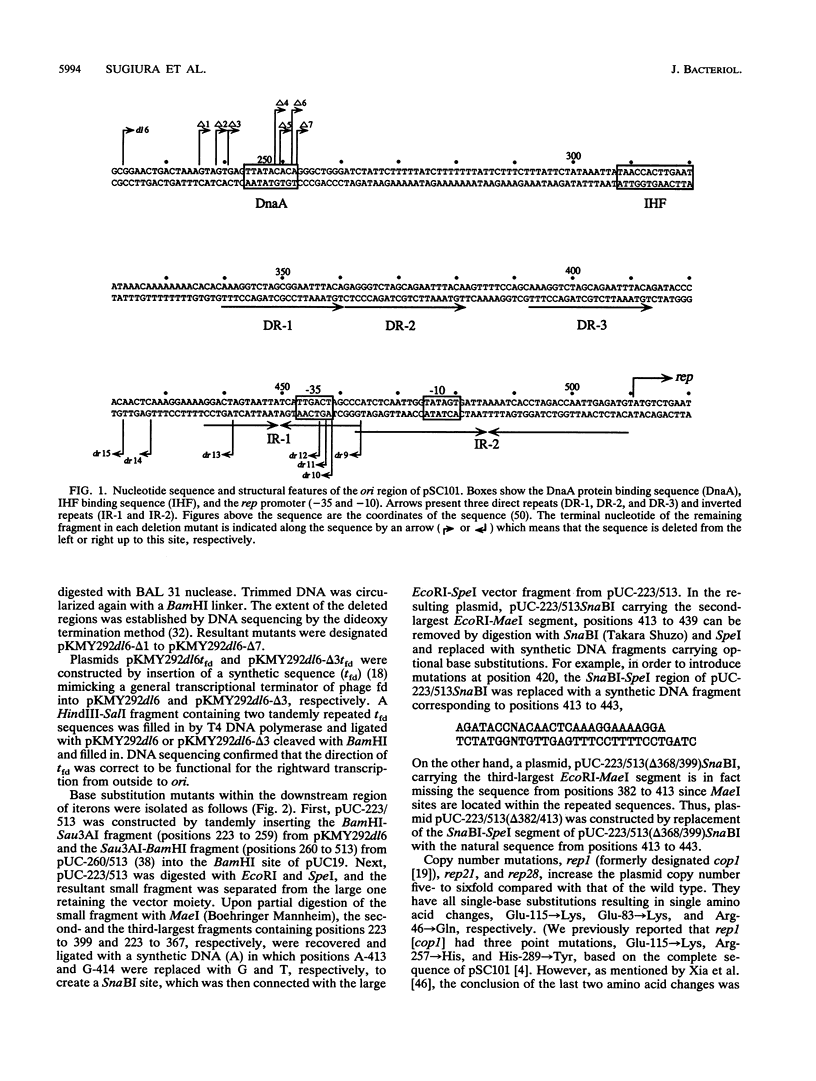
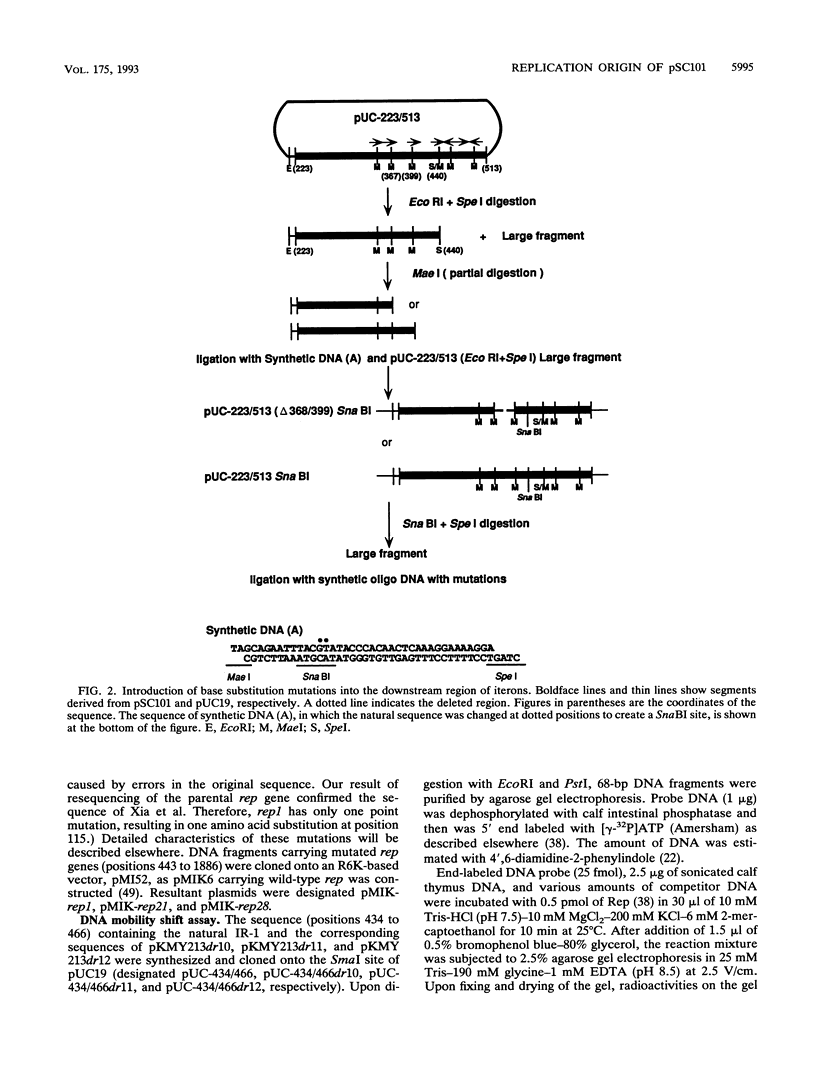
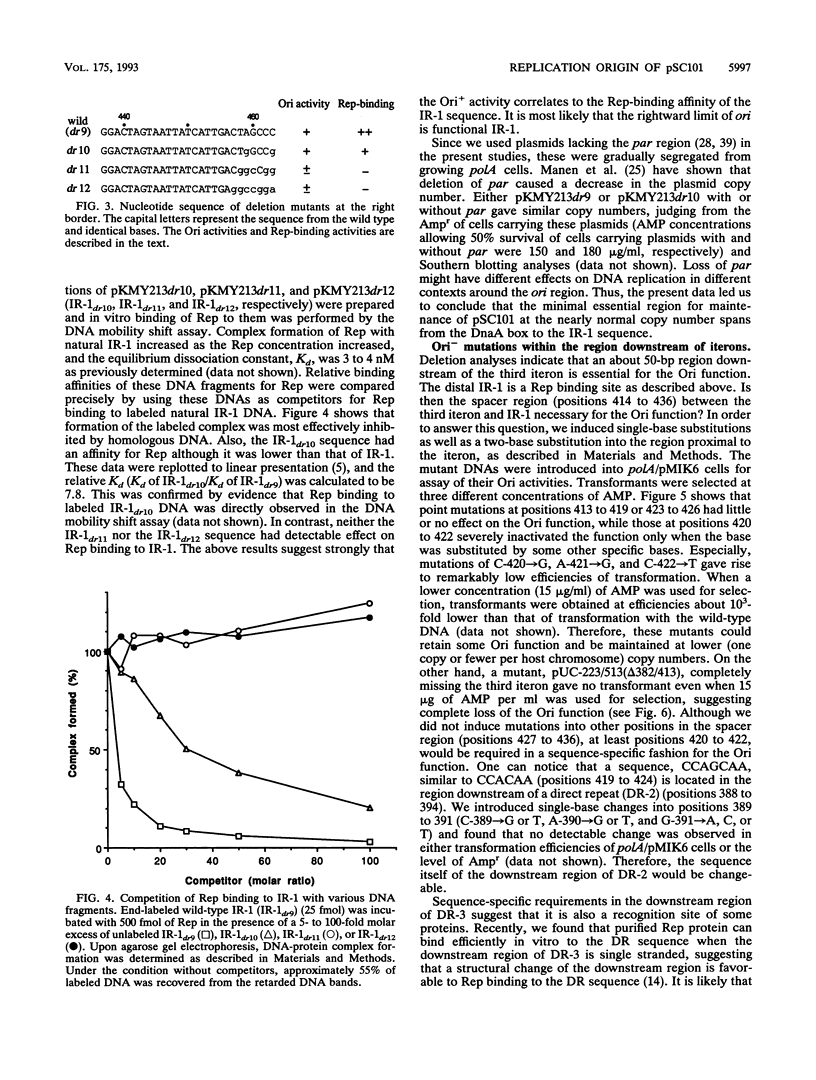
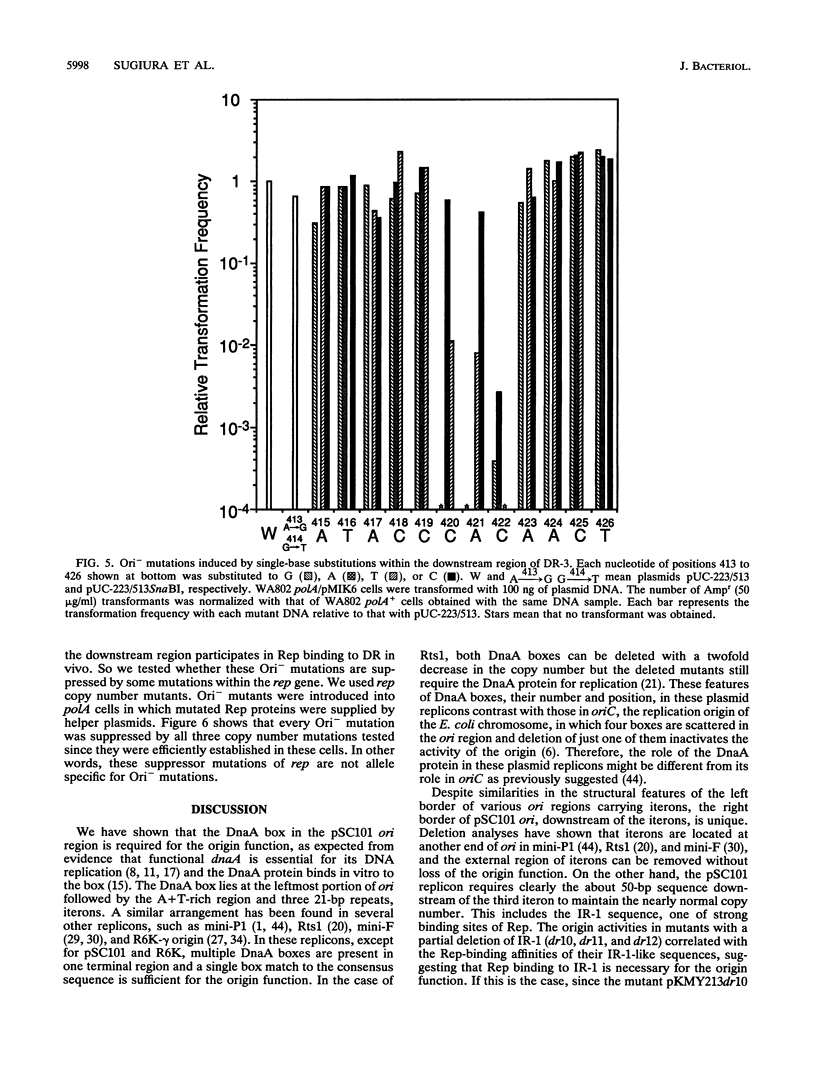
The minimal replication origin (ori) of the plasmid pSC101 was defined as an about 220-bp region under the condition that the Rep (or RepA) protein, a plasmid-encoded initiator protein, was supplied in trans. The DnaA box is located at one end of ori, as in other plasmids, like mini-F and P1. The other border is a strong binding site (IR-1) of Rep which is palindromic sequence and lies in an about 50-bp region beyond the repeated sequences (iterons) in ori. This IR-1 is located just upstream of another strong Rep binding site (IR-2), the operator site of the structure gene of Rep (rep), but its function has not been determined. The present study shows that the IR-1 sequence capable of binding to Rep is essential for plasmid replication with a nearly normal copy number. Furthermore, a region between the third iteron and IR-1 is also required in a sequence-specific fashion, since some one-base substitution in this region inactivate the origin function. It is likely that the region also is a recognition site of an unknown protein. Three copy number mutations of rep can suppress any one-base substitution mutation. On the other hand, the sequence of a spacer region between the second and the third iterons, which is similar to that of the downstream region of the third iteron, can be changed without loss of the origin function. The requirement of the region downstream of iterons in pSC101 seems to be unique among iteron-driven plasmid replicons.
Full text
PDF



Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Abeles A. L., Reaves L. D., Austin S. J. A single DnaA box is sufficient for initiation from the P1 plasmid origin. J Bacteriol. 1990 Aug;172(8):4386–4391. doi: 10.1128/jb.172.8.4386-4391.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abeles A. L., Snyder K. M., Chattoraj D. K. P1 plasmid replication: replicon structure. J Mol Biol. 1984 Mar 5;173(3):307–324. doi: 10.1016/0022-2836(84)90123-2. [DOI] [PubMed] [Google Scholar]
- Armstrong K. A., Acosta R., Ledner E., Machida Y., Pancotto M., McCormick M., Ohtsubo H., Ohtsubo E. A 37 X 10(3) molecular weight plasmid-encoded protein is required for replication and copy number control in the plasmid pSC101 and its temperature-sensitive derivative pHS1. J Mol Biol. 1984 May 25;175(3):331–348. doi: 10.1016/0022-2836(84)90352-8. [DOI] [PubMed] [Google Scholar]
- Bernardi A., Bernardi F. Complete sequence of pSC101. Nucleic Acids Res. 1984 Dec 21;12(24):9415–9426. doi: 10.1093/nar/12.24.9415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourgeois S., Riggs A. D. The lac repressor-operator interaction. IV. Assay and purification of operator DNA. Biochem Biophys Res Commun. 1970 Jan 23;38(2):348–354. doi: 10.1016/0006-291x(70)90719-9. [DOI] [PubMed] [Google Scholar]
- Bramhill D., Kornberg A. Duplex opening by dnaA protein at novel sequences in initiation of replication at the origin of the E. coli chromosome. Cell. 1988 Mar 11;52(5):743–755. doi: 10.1016/0092-8674(88)90412-6. [DOI] [PubMed] [Google Scholar]
- Churchward G., Linder P., Caro L. The nucleotide sequence of replication and maintenance functions encoded by plasmid pSC101. Nucleic Acids Res. 1983 Aug 25;11(16):5645–5659. doi: 10.1093/nar/11.16.5645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felton J., Wright A. Plasmid pSC101 replication in integratively suppressed cells requires dnaA function. Mol Gen Genet. 1979 Sep;175(2):231–233. doi: 10.1007/BF00425541. [DOI] [PubMed] [Google Scholar]
- Filutowicz M., Appelt K. The integration host factor of Escherichia coli binds to multiple sites at plasmid R6K gamma origin and is essential for replication. Nucleic Acids Res. 1988 May 11;16(9):3829–3843. doi: 10.1093/nar/16.9.3829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filutowicz M., Davis G., Greener A., Helinski D. R. Autorepressor properties of the pi-initiation protein encoded by plasmid R6K. Nucleic Acids Res. 1985 Jan 11;13(1):103–114. doi: 10.1093/nar/13.1.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frey J., Chandler M., Caro L. The effects of an Escherichia coli dnaAts mutation on the replication of the plasmids colE1 pSC101, R100.1 and RTF-TC. Mol Gen Genet. 1979 Jul 13;174(2):117–126. doi: 10.1007/BF00268349. [DOI] [PubMed] [Google Scholar]
- Friedman D. I. Integration host factor: a protein for all reasons. Cell. 1988 Nov 18;55(4):545–554. doi: 10.1016/0092-8674(88)90213-9. [DOI] [PubMed] [Google Scholar]
- Fueki T., Yamaguchi K. A single-stranded region located downstream of the repeated sequence in the ori region of pSC101 is required for binding of the Rep protein in vitro. J Biochem. 1991 Nov;110(5):775–779. doi: 10.1093/oxfordjournals.jbchem.a123658. [DOI] [PubMed] [Google Scholar]
- Fueki T., Yamaguchi K. Exonuclease III promotes in vitro binding of the replication initiator protein of plasmid pSC101 to the repeated sequences in the ori region. Biochim Biophys Acta. 1991 Jul 23;1089(3):325–330. doi: 10.1016/0167-4781(91)90172-i. [DOI] [PubMed] [Google Scholar]
- Fuller R. S., Funnell B. E., Kornberg A. The dnaA protein complex with the E. coli chromosomal replication origin (oriC) and other DNA sites. Cell. 1984 Oct;38(3):889–900. doi: 10.1016/0092-8674(84)90284-8. [DOI] [PubMed] [Google Scholar]
- Gamas P., Burger A. C., Churchward G., Caro L., Galas D., Chandler M. Replication of pSC101: effects of mutations in the E. coli DNA binding protein IHF. Mol Gen Genet. 1986 Jul;204(1):85–89. doi: 10.1007/BF00330192. [DOI] [PubMed] [Google Scholar]
- Hasunuma K., Sekiguchi M. Replication of plasmid pSC101 in Escherichia coli K12: requirement for dnaA function. Mol Gen Genet. 1977 Sep 9;154(3):225–230. doi: 10.1007/BF00571277. [DOI] [PubMed] [Google Scholar]
- Hirano M., Shigesada K., Imai M. Construction and characterization of plasmid and lambda phage vector systems for study of transcriptional control in Escherichia coli. Gene. 1987;57(1):89–99. doi: 10.1016/0378-1119(87)90180-6. [DOI] [PubMed] [Google Scholar]
- Itoh Y., Kamio Y., Terawaki Y. Essential DNA sequence for the replication of Rts1. J Bacteriol. 1987 Mar;169(3):1153–1160. doi: 10.1128/jb.169.3.1153-1160.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh Y., Terawaki Y. Replication properties of mini-Rts1 derivatives deleted for DnaA boxes in the replication origin. Plasmid. 1989 May;21(3):242–246. doi: 10.1016/0147-619x(89)90048-6. [DOI] [PubMed] [Google Scholar]
- Kapuściński J., Skoczylas B. Simple and rapid fluorimetric method for DNA microassay. Anal Biochem. 1977 Nov;83(1):252–257. doi: 10.1016/0003-2697(77)90533-4. [DOI] [PubMed] [Google Scholar]
- Linder P., Churchward G., Caro L. Plasmid pSC101 replication mutants generated by insertion of the transposon Tn1000. J Mol Biol. 1983 Oct 25;170(2):287–303. doi: 10.1016/s0022-2836(83)80149-1. [DOI] [PubMed] [Google Scholar]
- Linder P., Churchward G., Xia G. X., Yu Y. Y., Caro L. An essential replication gene, repA, of plasmid pSC101 is autoregulated. J Mol Biol. 1985 Feb 5;181(3):383–393. doi: 10.1016/0022-2836(85)90227-x. [DOI] [PubMed] [Google Scholar]
- Manen D., Goebel T., Caro L. The par region of pSC101 affects plasmid copy number as well as stability. Mol Microbiol. 1990 Nov;4(11):1839–1846. doi: 10.1111/j.1365-2958.1990.tb02032.x. [DOI] [PubMed] [Google Scholar]
- Manen D., Upegui-Gonzalez L. C., Caro L. Monomers and dimers of the RepA protein in plasmid pSC101 replication: domains in RepA. Proc Natl Acad Sci U S A. 1992 Oct 1;89(19):8923–8927. doi: 10.1073/pnas.89.19.8923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McEachern M. J., Filutowicz M., Helinski D. R. Mutations in direct repeat sequences and in a conserved sequence adjacent to the repeats result in a defective replication origin in plasmid R6K. Proc Natl Acad Sci U S A. 1985 Mar;82(5):1480–1484. doi: 10.1073/pnas.82.5.1480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meacock P. A., Cohen S. N. Partitioning of bacterial plasmids during cell division: a cis-acting locus that accomplishes stable plasmid inheritance. Cell. 1980 Jun;20(2):529–542. doi: 10.1016/0092-8674(80)90639-x. [DOI] [PubMed] [Google Scholar]
- Murakami Y., Ohmori H., Yura T., Nagata T. Requirement of the Escherichia coli dnaA gene function for ori-2-dependent mini-F plasmid replication. J Bacteriol. 1987 Apr;169(4):1724–1730. doi: 10.1128/jb.169.4.1724-1730.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murotsu T., Tsutsui H., Matsubara K. Identification of the minimal essential region for the replication origin of miniF plasmid. Mol Gen Genet. 1984;196(2):373–378. doi: 10.1007/BF00328075. [DOI] [PubMed] [Google Scholar]
- Nordström K. Control of plasmid replication--how do DNA iterons set the replication frequency? Cell. 1990 Dec 21;63(6):1121–1124. doi: 10.1016/0092-8674(90)90405-4. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaefer C., Messer W. DnaA protein/DNA interaction. Modulation of the recognition sequence. Mol Gen Genet. 1991 Apr;226(1-2):34–40. doi: 10.1007/BF00273584. [DOI] [PubMed] [Google Scholar]
- Stalker D. M., Kolter R., Helinski D. R. Nucleotide sequence of the region of an origin of replication of the antibiotic resistance plasmid R6K. Proc Natl Acad Sci U S A. 1979 Mar;76(3):1150–1154. doi: 10.1073/pnas.76.3.1150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenzel T. T., MacAllister T., Bastia D. Cooperativity at a distance promoted by the combined action of two replication initiator proteins and a DNA bending protein at the replication origin of pSC101. Genes Dev. 1991 Aug;5(8):1453–1463. doi: 10.1101/gad.5.8.1453. [DOI] [PubMed] [Google Scholar]
- Stenzel T. T., Patel P., Bastia D. The integration host factor of Escherichia coli binds to bent DNA at the origin of replication of the plasmid pSC101. Cell. 1987 Jun 5;49(5):709–717. doi: 10.1016/0092-8674(87)90547-2. [DOI] [PubMed] [Google Scholar]
- Sugiura S., Nakatani S., Mizukami Y., Hase T., Hirokawa H., Masamune Y. Characterization of a mini plasmid isolated from Shigella sonnei. J Biochem. 1984 Oct;96(4):1193–1204. doi: 10.1093/oxfordjournals.jbchem.a134937. [DOI] [PubMed] [Google Scholar]
- Sugiura S., Tanaka M., Masamune Y., Yamaguchi K. DNA binding properties of purified replication initiator protein (Rep) encoded by plasmid pSC101. J Biochem. 1990 Mar;107(3):369–376. doi: 10.1093/oxfordjournals.jbchem.a123052. [DOI] [PubMed] [Google Scholar]
- Tucker W. T., Miller C. A., Cohen S. N. Structural and functional analysis of the par region of the pSC 10 1 plasmid. Cell. 1984 Aug;38(1):191–201. doi: 10.1016/0092-8674(84)90540-3. [DOI] [PubMed] [Google Scholar]
- Vocke C., Bastia D. DNA-protein interaction at the origin of DNA replication of the plasmid pSC101. Cell. 1983 Dec;35(2 Pt 1):495–502. doi: 10.1016/0092-8674(83)90183-6. [DOI] [PubMed] [Google Scholar]
- Vocke C., Bastia D. Primary structure of the essential replicon of the plasmid pSC101. Proc Natl Acad Sci U S A. 1983 Nov;80(21):6557–6561. doi: 10.1073/pnas.80.21.6557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vocke C., Bastia D. The replication initiator protein of plasmid pSC101 is a transcriptional repressor of its own cistron. Proc Natl Acad Sci U S A. 1985 Apr;82(8):2252–2256. doi: 10.1073/pnas.82.8.2252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahle E., Kornberg A. The partition locus of plasmid pSC101 is a specific binding site for DNA gyrase. EMBO J. 1988 Jun;7(6):1889–1895. doi: 10.1002/j.1460-2075.1988.tb03022.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickner S., Hoskins J., Chattoraj D., McKenney K. Deletion analysis of the mini-P1 plasmid origin of replication and the role of Escherichia coli DnaA protein. J Biol Chem. 1990 Jul 15;265(20):11622–11627. [PubMed] [Google Scholar]
- Wickner S., Hoskins J., McKenney K. Monomerization of RepA dimers by heat shock proteins activates binding to DNA replication origin. Proc Natl Acad Sci U S A. 1991 Sep 15;88(18):7903–7907. doi: 10.1073/pnas.88.18.7903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia G. X., Manen D., Goebel T., Linder P., Churchward G., Caro L. A copy-number mutant of plasmid pSC101. Mol Microbiol. 1991 Mar;5(3):631–640. doi: 10.1111/j.1365-2958.1991.tb00734.x. [DOI] [PubMed] [Google Scholar]
- Yamaguchi K., Masamune Y. Autogenous regulation of synthesis of the replication protein in plasmid pSC101. Mol Gen Genet. 1985;200(3):362–367. doi: 10.1007/BF00425718. [DOI] [PubMed] [Google Scholar]
- Yamaguchi K., Tomizawa J. Establishment of Escherichia coli cells with an integrated high copy number plasmid. Mol Gen Genet. 1980;178(3):525–533. doi: 10.1007/BF00337857. [DOI] [PubMed] [Google Scholar]
- Yamaguchi K., Yamaguchi M. The replication origin of pSC101: the nucleotide sequence and replication functions of the ori region. Gene. 1984 Jul-Aug;29(1-2):211–219. doi: 10.1016/0378-1119(84)90181-1. [DOI] [PubMed] [Google Scholar]
- Yanisch-Perron C., Vieira J., Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33(1):103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]


