Abstract
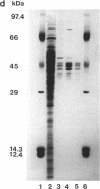
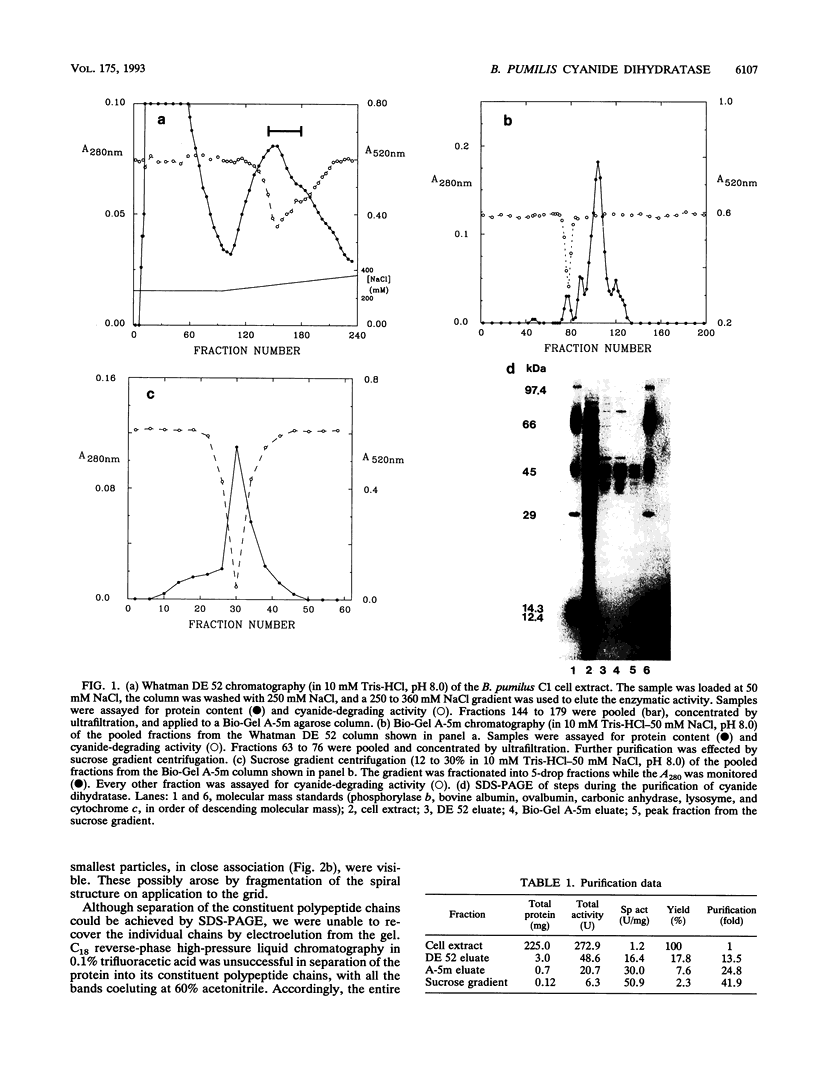
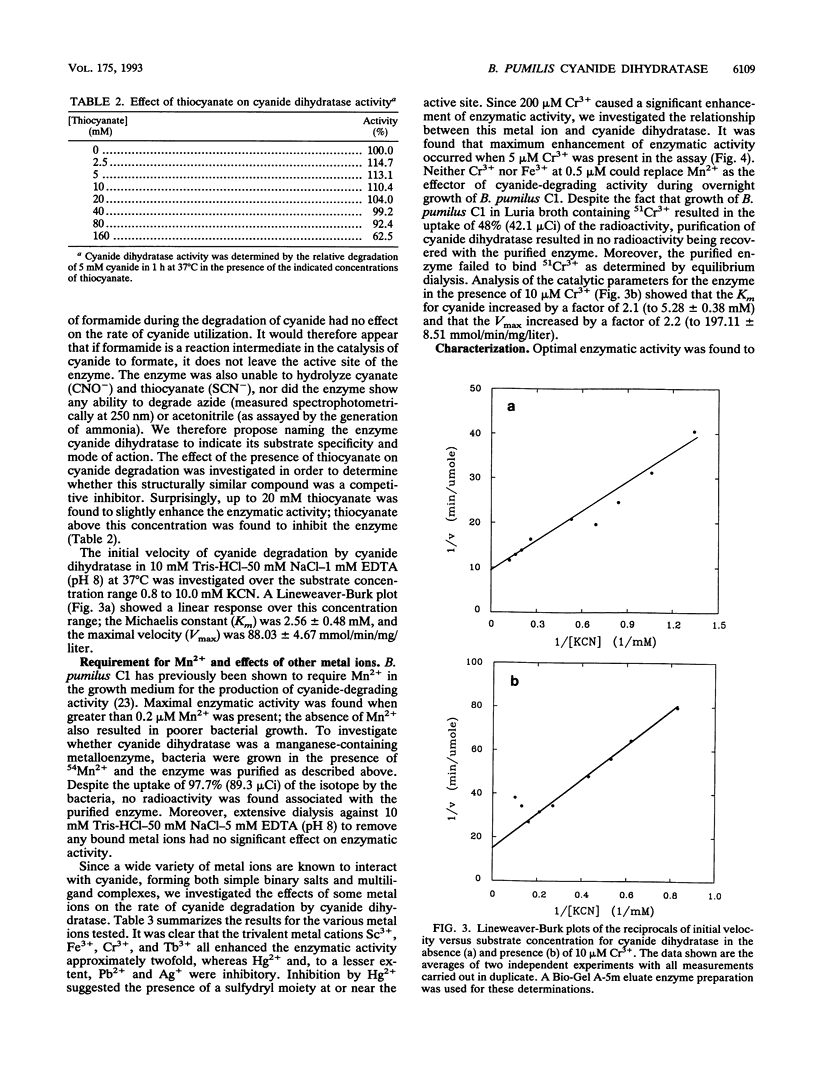
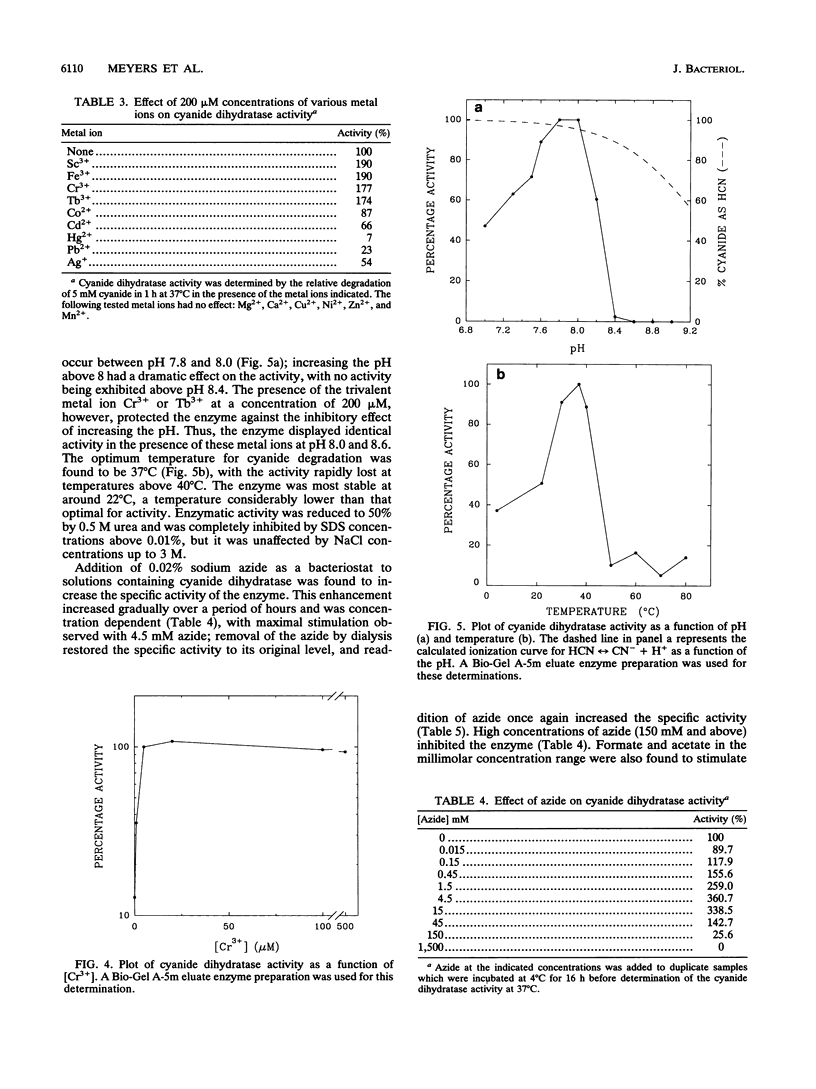
A cyanide-degrading enzyme from Bacillus pumilus C1 has been purified and characterized. This enzyme consisted of three polypeptides of 45.6, 44.6, and 41.2 kDa; the molecular mass by gel filtration was 417 kDa. Electron microscopy revealed a multimeric, rod-shaped protein approximately 9 by 50 nm. Cyanide was rapidly degraded to formate and ammonia. Enzyme activity was optimal at 37 degrees C and pH 7.8 to 8.0. Activity was enhanced by Sc3+, Cr3+, Fe3+, and Tb3+; enhancement was independent of metal ion concentration at concentrations above 5 microM. Reversible enhancement of enzymatic activity by azide was maximal at 4.5 mM azide and increased with time. No activity was recorded with the cyanide substrate analogs CNO-, SCN-, CH3CN, and N3- and the possible degradation intermediate HCONH2. Kinetic studies indicated a Km of 2.56 +/- 0.48 mM for cyanide and a Vmax of 88.03 +/- 4.67 mmol of cyanide per min/mg/liter. The Km increased approximately twofold in the presence of 10 microM Cr3+ to 5.28 +/- 0.38 mM for cyanide, and the Vmax increased to 197.11 +/- 8.51 mmol of cyanide per min/mg/liter. We propose naming this enzyme cyanide dihydratase.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- FAWCETT J. K., SCOTT J. E. A rapid and precise method for the determination of urea. J Clin Pathol. 1960 Mar;13:156–159. doi: 10.1136/jcp.13.2.156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fry W. E., Millar R. L. Cyanide degradion by an enzyme from Stemphylium loti. Arch Biochem Biophys. 1972 Aug;151(2):468–474. doi: 10.1016/0003-9861(72)90523-1. [DOI] [PubMed] [Google Scholar]
- Harris R., Knowles C. J. Isolation and growth of a Pseudomonas species that utilizes cyanide as a source of nitrogen. J Gen Microbiol. 1983 Apr;129(4):1005–1011. doi: 10.1099/00221287-129-4-1005. [DOI] [PubMed] [Google Scholar]
- Ingvorsen K., Højer-Pedersen B., Godtfredsen S. E. Novel cyanide-hydrolyzing enzyme from Alcaligenes xylosoxidans subsp. denitrificans. Appl Environ Microbiol. 1991 Jun;57(6):1783–1789. doi: 10.1128/aem.57.6.1783-1789.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knowles C. J., Bunch A. W. Microbial cyanide metabolism. Adv Microb Physiol. 1986;27:73–111. doi: 10.1016/s0065-2911(08)60304-5. [DOI] [PubMed] [Google Scholar]
- Knowles C. J. Microorganisms and cyanide. Bacteriol Rev. 1976 Sep;40(3):652–680. doi: 10.1128/br.40.3.652-680.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunz D. A., Nagappan O., Silva-Avalos J., Delong G. T. Utilization of cyanide as nitrogenous substrate by Pseudomonas fluorescens NCIMB 11764: evidence for multiple pathways of metabolic conversion. Appl Environ Microbiol. 1992 Jun;58(6):2022–2029. doi: 10.1128/aem.58.6.2022-2029.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Meyers P. R., Gokool P., Rawlings D. E., Woods D. R. An efficient cyanide-degrading Bacillus pumilus strain. J Gen Microbiol. 1991 Jun;137(6):1397–1400. doi: 10.1099/00221287-137-6-1397. [DOI] [PubMed] [Google Scholar]
- Skowronski B., Strobel G. A. Cyanide resistance and cyanide utilization by a strain of Bacillus pumilus. Can J Microbiol. 1969 Jan;15(1):93–98. doi: 10.1139/m69-014. [DOI] [PubMed] [Google Scholar]
- Wang P., Matthews D. E., VanEtten H. D. Purification and characterization of cyanide hydratase from the phytopathogenic fungus Gloeocercospora sorghi. Arch Biochem Biophys. 1992 Nov 1;298(2):569–575. doi: 10.1016/0003-9861(92)90451-2. [DOI] [PubMed] [Google Scholar]
- Wang P., VanEtten H. D. Cloning and properties of a cyanide hydratase gene from the phytopathogenic fungus Gloeocercospora sorghi. Biochem Biophys Res Commun. 1992 Sep 16;187(2):1048–1054. doi: 10.1016/0006-291x(92)91303-8. [DOI] [PubMed] [Google Scholar]