Abstract
The locus coeruleus (LC) is a major noradrenergic brain nucleus that regulates states of arousal, optimizes task-oriented decision-making, and may also play an important role in modulating the activity of the reproductive neuroendocrine axis. Rodent studies have shown that the LC is responsive to glutamate receptor agonists, and that it expresses various glutamate receptor subunits. However, glutamate receptor subunit expression has not been extensively examined in the primate LC. We previously demonstrated expression of the NR1 NMDA glutamate receptor subunit in the rhesus macaque LC, and now extend this work by also examining the expression of non-NMDA (AMPA and kainate) ionotropic glutamate receptor subunits. Using in situ hybridization histochemistry and immunohistochemistry, we confirmed the presence of the obligatory NR1 subunit in the LC. In addition, we demonstrated expression of the AMPA glutamate receptor subunits GluR1, GluR2 and GluR3. More extensive receptor profiling, using rhesus monkey gene microarrays (Affymetrix GeneChip®), further corroborated the histological findings and showed expression of mRNA encoding ionotropic glutamate receptor subunits NR2A, NR2D, GluR4, and GluR6, as well as the metabotropic glutamate receptor subunits mGluR1, mGluR3, mGluR4, mGluR5 and mGluR7. These data provide a foundation for future examination of how changes in glutamate receptor composition contribute to the control of primate physiology.
Keywords: Locus coeruleus, Microarray, Gene array, Rhesus macaque, Glutamate receptor, in situ hybridization
1. Introduction
The locus coeruleus (LC) is the principal noradrenergic nucleus of the brain and is connected to areas as wide ranging as the cerebral cortex, thalamus, hypothalamus, olfactory bulb, cerebellum, midbrain and spinal cord (Moore and Bloom, 1979; Foote et al., 1983; Loughlin et al., 1986; Luppi et al., 1995; Berridge and Waterhouse, 2003). Early research implicated LC activity in a range of physiological responses originally described in terms of “arousal” and responses to “stressful” stimuli (Cedarbaum and Aghajanian, 1978; Aston-Jones and Bloom, 1981; Elam et al., 1985; Abercrombie and Jacobs, 1987; Curtis et al., 1993; Rasmussen, 1995; Singewald and Philippu, 1998). More recently, drawing extensively from primate studies (Clayton et al., 2004; Rajkowski et al., 2004), Aston-Jones has proposed an integrative “adaptive gain” hypothesis interpreting the LC as cognitive “attention filter” to optimize performance based on the outcome of task-related decision processes (Aston-Jones and Cohen, 2005a; 2005b). Overall, the LC integrates information regarding cognition, states of awareness, alertness and associated behavior (Berridge and Waterhouse, 2003). Related to such integration, is LC regulation of sleep (Cirelli et al., 1996; Gonzalez et al., 1996; Aston-Jones, 2005) and circadian rhythms (Gonzalez and Aston-Jones, 2006). For example, the LC acts in concert with hypocretin/orexin (Horvath et al., 1999; Bourgin et al., 2000; Espana et al., 2005; Downs et al., 2007) and is a key player in awareness/alertness disorders such as narcolepsy (Fruhstorfer et al., 1989; Wu et al., 1999).
In addition, LC neural connections to the hypothalamus (Mason and Fibiger, 1979; Sawchenko and Swanson, 1981; Sawchenko and Swanson, 1982; Sawchenko et al., 1985; Loughlin et al., 1986; Cunningham and Sawchenko, 1988) provide an anatomical basis for LC function in mammalian reproduction. Detailed work in rabbits (induced ovulators) show that LC-modulated norepinephrine (NE) and NE transporter (NET) levels regulate hypothalamic gonadotropin releasing hormone (GnRH) and luteinizing hormone (LH) in response to coitus or estradiol stimulation (Yang et al., 1997; Yang et al., 1997; Pau et al., 1998). These studies support the hypothesis that the LC is a relay station (Pau et al., 1997) for reproductive signals reaching hypothalamic GnRH neurons in a proposed estrogen-induced NE→GnRH→LH-surge mechanism (Spies et al., 1997; Yang et al., 1998) and suggest a mechanism by which the LC may be involved in hypothalamo-pituitary-gonadal (HPG) regulatory feedback. An LC-mediated HPG relay involving NE may also exist in primates, and be involved in the generation of GnRH-regulated pulsatile LH release (Terasawa et al., 1988; Pau et al., 1989; Gearing and Terasawa, 1991) as well as the LH surge (Pau et al., 2000).
The primate reproductive neuroendocrine axis is also profoundly influenced by glutamatergic pathways. These have been shown to activate GnRH release and to advance the onset of puberty (Wilson and Knobil, 1982; Gay and Plant, 1988; Plant et al., 1989; Reyes et al., 1991; Medhamurthy et al., 1992). However, because GnRH neurons show only low levels of glutamate receptor expression, non-hypothalamic glutamate-responsive noradrenergic afferents may play an important intermediary role (Leranth et al., 1988; Urbanski et al., 1996). The goal of the present study was to add credence to this possible mechanism, by examining the expression of various glutamate receptor subunits in the primate LC.
The neurotransmitter glutamate is highly abundant in the mammalian brain (Brann, 1995; Brann and Mahesh, 1995) and appears to be involved in most LC functions. For example, there is a wealth of data showing that noxious or stressful stimuli can stimulate glutamate release (likely from afferent neurons) in the LC (Singewald et al., 1994; Singewald et al., 1995; Singewald et al., 1996), and that the responses of the LC to activated afferents from the prefrontal cortex are mediated through glutamate receptors (Jodo and Aston-Jones, 1997). Such glutamatergic activation of the LC influences efferent synaptic responses involved in neural plasticity and memory (Walling and Harley, 2004).
Ionotropic glutamate receptors (ligand-gated ion-channels) belong to three pharmacologically defined classes named after selected agonists: N-methyl-D-aspartate (NMDA), α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid (AMPA), and kainate (Gasic and Hollmann, 1992; Hollmann and Heinemann, 1994; Dingledine et al., 1999). We have begun characterizing the glutamate receptor composition of the rhesus macaque LC by demonstrating of the presence of NR1 (i.e., NMDAR1), the obligatory glutamate receptor subunit (Urbanski et al., 1997). Our primary goal in the current study was to extend this research by providing a more comprehensive characterization of glutamate receptor subunit expression in the rhesus macaque LC. The recent completion of genome sequencing in this nonhuman primate, and the demonstration of 97.5% identity between the coding regions of macaques and humans (Gibbs et al., 2007), make the interpretation of glutamate receptor function in the macaque LC highly relevant to hypotheses regarding human brain physiology.
2. Results
2.1 LC localization
The LC was identified in coronal brain sections using immunohistochemistry for tyrosine hydroxylase (TH), a key enzyme in the biosynthesis of catecholamines (Fig. 1). As expected, the expression level of TH was characteristically high and concentrated in a discrete elongated bilateral nucleus, immediately lateral to the fourth ventricle and medial to the superior cerebellar peduncle. Nissl staining showed the mensencephalic trigeminal nerve tract laterally abutting the LC.
Figure 1.
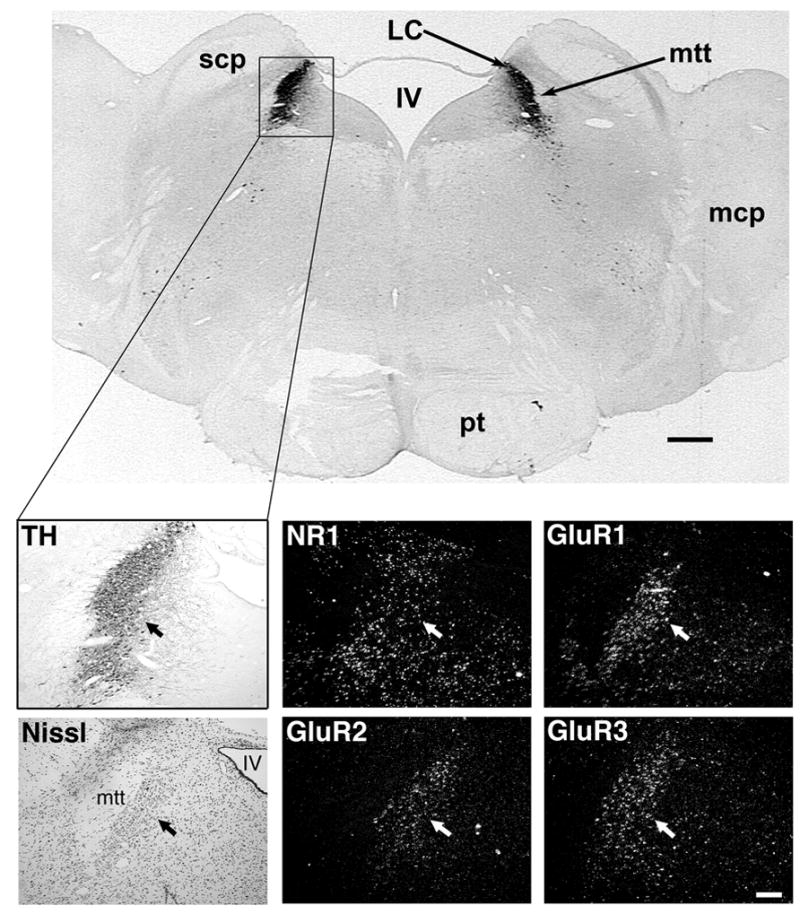
Localization of glutamate receptor subunit gene expression in the rhesus macaque LC, as revealed by in situ hybridization. The top panel shows a frontal section of the hindbrain immunostained for TH: LC = locus coeruleus, scp = superior cerebellar peduncle, mtt = mesencephalic trigeminal nerve tract, IV = fourth ventricle, mcp = medial cerebellar peduncle, pt = pyramidal tract. Black scale bar = 1mm.
The lower six panels represent adjacent coronal 25-μm sections. The light panels show landmarks used to localize the LC. TH = immunostaining for TH using DAB as the chromogen. Nissl = cresyl violet stain for Nissl substance. The TH-positive neurons of the LC (arrows) lie between the fourth ventricle (IV) and the mesencephalic trigeminal nerve tract (mtt); Dark panels show in situ hybridization for glutamate receptor subunits NMDAR1 (NR1), GluR1, GluR2 and GluR3 in the LC. White scale bar = 250 μm.
2.2 In situ hybridization and immunohistochemistry
Probes used for in situ hybridization (performed on sections adjacent to those showing TH immunoreactivity) revealed high binding to the NR1 NMDA receptor subunit mRNA in the LC and in the surrounding tissue (Fig. 1). Similar results were obtained using probes for the GluR1, GluR2 and GluR3 AMPA receptor subunit mRNA (Fig. 1).
On a separate set of sections (also adjacent to TH immunopositive sections), immunohistochemistry using antibodies against NR1, GluR1 and GluR2/3 disclosed glutamate receptor subunit protein expression in LC neural tissue (Fig. 2). These immunopositive perikarya had a similar distribution pattern to TH within the LC, were present in approximately the same number and had similar perikaryal size.
Figure 2.
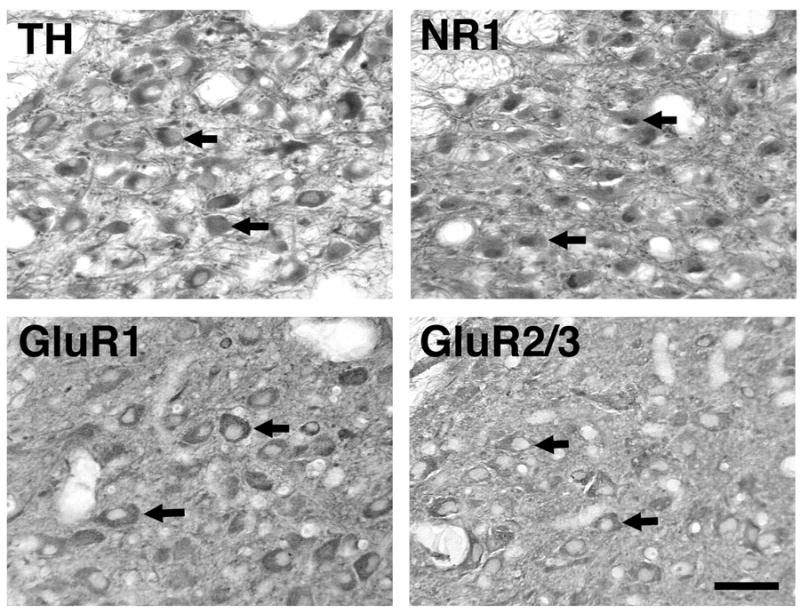
Distribution of TH and glutamate receptor subunit proteins in individual cell bodies of the rhesus macaque LC, as revealed by immunohistochemistry using DAB as the chromogen. Arrows indicate representative cell bodies showing positive immunostaining. Scale bar = 100 μm.
2.3 Gene profiling using cDNA microarrays
Because of limited availablilty of specific probes and antibodies, we also attempted a more comprehensive assessment of LC glutamate receptor subunit expression using microarray gene analysis. The results demonstrated TH mRNA in tissue samples from both of the rhesus macaques examined in the current study, thereby confirming that the RNA was derived correctly from the LC. The absence of glial fibrillary acidic protein (GFAP) expression (Table 1), used as a gene marker for astrocytes, corroborated that RNA contamination from astrocyte-rich pericoerulear tissue was low. Although the primary focus of the gene microarray analysis was on glutamate receptor subunits, we also examined whether the LC has the capacity to produce the neurotransmitter glutamate itself. The analysis showed that each of the three common vesicular glutamate transporter genes (VGLUT1, VGLUT2, and VGLUT3) was expressed in the LC, with VGLUT2 being particularly abundant (Table 1). In addition, genes encoding glutaminase (the enzyme that catalyzes the conversion of glutamine to glutamate) and glutamic acid decarboxylase (the enzyme that catalyzes the conversion of glutamate to GABA) were also highly expressed in the LC of both animals (Table 1).
Table 1.
Location-specific expression of vesicular glutamate transporter and housekeeping genes in the rhesus macaque locus coeruleus
| Affymetrix Probe Set ID | Common Gene Description | Gene Symbol | Animal 1 | Animal 2 |
|---|---|---|---|---|
| MmugDNA.5125.1.S1_at | Tyrosine hydroxylase | TH | +++ | + |
| MmugDNA.37177.1.S1_at | Glial fibrillary acidic protein | GFAP | − | − |
| MmugDNA.6553.1.S1_at | Solute carrier family 17 (sodium-dependent inorganic phosphate cotransporter), member 6 (VGLUT2) | SLC17A6 | +++ | +++ |
| MmugDNA.26376.1.S1_at | Solute carrier family 17 (sodium-dependent inorganic phosphate cotransporter), member 7 (VGLUT1) | SLC17A7 | + | ++ |
| MmuSTS.3832.1.S1_at | Solute carrier family 17 (sodium-dependent inorganic phosphate cotransporter), member 8 (VGLUT3) | SLC17A8 | + | + |
| Mmu.7638.1.S1_at | Glutaminase C | GLS | + | + |
| MmugDNA.10260.1.S1_at | Glutaminase C | GLS | +++ | +++ |
| MmugDNA.12940.1.S1_at | Glutaminase C | GLS | ++ | ++ |
| MmugDNA.15005.1.S1_at | Glutaminase C | GLS | + | ++ |
| MmugDNA.31518.1.S1_at | Glutaminase C | GLS | − | − |
| MmuAffx.22.1.S1_at | Glutamate decarboxylase 1 | GAD1 | − | + |
| MmugDNA.20165.1.S1_at | Glutamate decarboxylase 1 | GAD1 | + | + |
| MmugDNA.40699.1.S1_at | Glutamate decarboxylase 1 | GAD1 | +++ | +++ |
| MmugDNA.3230.1.S1_at | Glutamate decarboxylase 2 | GAD2 | − | + |
| MmugDNA.40703.1.S1_at | Glutamate decarboxylase 2 | GAD2 | +++ | +++ |
| MmugDNA.6977.1.S1_at | Glutamate decarboxylase 2 | GAD2 | − | − |
| MmugDNA.28776.1.S1_s_at | Actin, beta | ACTB | +++ | +++ |
| MmugDNA.34587.1.S1_s_at | Glyceraldehyde-3-phosphate dehydrogenase | GAPDH | +++ | +++ |
Table legend: “+” and “−” symbols represent values obtained using Affymetrix GCOS analysis, and global scaling to an average target intensity of 200. (−) = “undetectable”, (+/−) = “marginally present”, (+) = “signal intensity < 500”, (++) = “signal intensity 500–1000”, (+++) = “signal intensity >1000”.
Detection levels for housekeeping genes β-actin (ACTB) and glyceraldehyde 3-phosphate dehydrogenase (GAPDH) were high and comparable between the two animals (Table 1). Signal intensities for ACTB differed between the two animals by less that 4%, and the intensity of the GAPDH signal was three to four times that of the ACTB signal (data not shown).
Microarray analysis of the ionotropic glutamate receptor subunit sequences in the LC (Table 2) showed mRNA expression encoding NR1, NR2A and NR2D NMDA receptor subunits, as well as GluR1, GluR2, GluR3 and GluR4 AMPA receptor subunits. Expression of mRNA for GluR2 and 4 was particularly strong, while expression for GluR3 was particularly weak, with convincing chip hybridization demonstrated in only one of four GluR3 probe sets. Of the kainate receptor subunit sequences, only a low level GluR6 mRNA expression was evident. No mRNA for the delta subunits appeared to be expressed in the LC. Microarray analysis for the metabotropic glutamate receptor subunit sequences (Table 3) showed expression of mGluR1, mGluR3, mGluR4, mGluR5 and mGluR7, but not mGluR2, mGluR6 and mGluR8 mRNA in the LC.
Table 2.
Expression of ionotropic glutamate receptor subunit mRNA in the rhesus macaque locus coeruleus
| Affymetrix Probe Set ID | Common Gene Description | Gene Symbol | Animal 1 | Animal 2 |
|---|---|---|---|---|
| MmugDNA.31018.1.S1_at | Glutamate Receptor (zeta 1), Ionotropic (NMDAR1)/(NR1) | Grin1 | + | ++ |
| MmugDNA.31300.1.S1_s_at | Glutamate Receptor (zeta 1), Ionotropic (NMDAR1)/(NR1) | Grin1 | − | − |
| MmugDNA.36312.1.S1_s_at | Glutamate Receptor (zeta 1), Ionotropic (NMDAR1)/(NR1) | Grin1 | − | − |
| MmugDNA.26473.1.S1_at | Glutamate Receptor (epsilon 1), Ionotropic (NMDAR2A)/(NR2A) | Grin2A | ++ | ++ |
| MmugDNA.42589.1.S1_at | Glutamate Receptor (epsilon 1), Ionotropic (NMDAR2A)/(NR2A) | Grin2A | + | + |
| MmuSTS.1.1.S1_at | Glutamate Receptor (epsilon 1), Ionotropic (NMDAR2A)/(NR2A) | Grin2A | + | ++ |
| MmugDNA.38537.1.S1_at | Glutamate Receptor (epsilon 2), Ionotropic (NMDAR2B)/(NR2B) | Grin2B | − | − |
| MmugDNA.12768.1.S1_at | Glutamate Receptor (epsilon 3), Ionotropic (NMDAR2C)/(NR2C) | Grin2C | − | +/− |
| MmugDNA.2167.1.S1_at | Glutamate Receptor (epsilon 3), Ionotropic (NMDAR2C)/(NR2C) | Grin2C | − | + |
| MmugDNA.37367.1.S1_at | Glutamate Receptor (epsilon 3), Ionotropic (NMDAR2C)/(NR2C) | Grin2C | − | − |
| MmugDNA.9830.1.S1_at | Glutamate Receptor (epsilon 4), Ionotropic (NMDAR2D)/(NR2D) | Grin2D | + | + |
| MmugDNA.35584.1.S1_at | Glutamate Receptor, Ionotropic (NMDAR3A)/(NR3A) | Grin3A | − | − |
| MmugDNA.43206.1.S1_at | Glutamate Receptor, Ionotropic (NMDAR3B)/(NR3B) | Grin3B | − | − |
| MmugDNA.13803.1.S1_at | Glutamate Receptor 1, Ionotropic (GluR1)/(AMPA1) | Gria1 | +/− | − |
| MmugDNA.20338.1.S1_at | Glutamate Receptor 1, Ionotropic (GluR1)/(AMPA1) | Gria1 | + | ++ |
| MmugDNA.14406.1.S1_at | Glutamate Receptor 2, Ionotropic (GluR2)/(AMPA2) | Gria2 | − | − |
| MmugDNA.27764.1.S1_at | Glutamate Receptor 2, Ionotropic (GluR2)/(AMPA2) | Gria2 | +++ | +++ |
| MmugDNA.37346.1.S1_at | Glutamate Receptor 2, Ionotropic (GluR2)/(AMPA2) | Gria2 | +++ | +++ |
| MmugDNA.16285.1.S1_at | Glutamate Receptor 3, Ionotropic (GluR3)/(AMPA3) | Gria3 | − | − |
| MmugDNA.16285.1.S1_s_at | Glutamate Receptor 3, Ionotropic (GluR3)/(AMPA3) | Gria3 | − | + |
| MmugDNA.17786.1.S1_at | Glutamate Receptor 3, Ionotropic (GluR3)/(AMPA3) | Gria3 | − | − |
| MmugDNA.32084.1.S1_at | Glutamate Receptor 3, Ionotropic (GluR3)/(AMPA3) | Gria3 | + | + |
| MmugDNA.20418.1.S1_at | Glutamate Receptor 4, Ionotropic (GluR4)/(AMPA4) | Gria4 | + | + |
| MmugDNA.29490.1.S1_at | Glutamate Receptor 4, Ionotropic (GluR4)/(AMPA4) | Gria4 | − | + |
| MmugDNA.35630.1.S1_at | Glutamate Receptor 4, Ionotropic (GluR4)/(AMPA4) | Gria4 | − | − |
| MmugDNA.7637.1.S1_at | Glutamate Receptor 4, Ionotropic (GluR4)/(AMPA4) | Gria4 | +++ | +++ |
| MmugDNA.16287.1.S1_at | Glutamate Receptor 5, Ionotropic (GluR5)/(Kainate 1) | Grik1 | − | − |
| MmugDNA.5312.1.S1_at | Glutamate Receptor 5, Ionotropic (GluR5)/(Kainate 1) | Grik1 | − | − |
| MmugDNA.23700.1.S1_at | Glutamate Receptor 6, Ionotropic (GluR6)/(Kainate 2) | Grik2 | + | + |
| MmugDNA.27611.1.S1_at | Glutamate Receptor 6, Ionotropic (GluR6)/(Kainate 2) | Grik2 | − | − |
| MmugDNA.35699.1.S1_at | Glutamate Receptor 6, Ionotropic (GluR6)/(Kainate 2) | Grik2 | + | + |
| MmugDNA.37030.1.S1_at | Glutamate Receptor 6, Ionotropic (GluR6)/(Kainate 2) | Grik2 | − | − |
| MmugDNA.42517.1.S1_at | Glutamate Receptor 6, Ionotropic (GluR6)/(Kainate 2) | Grik2 | − | − |
| MmugDNA.8334.1.S1_at | Glutamate Receptor 6, Ionotropic (GluR6)/(Kainate 2) | Grik2 | + | + |
| MmuSTS.4845.1.S1_at | Glutamate Receptor 7, Ionotropic (GluR7)/(Kainate 3) | Grik3 | − | − |
| MmugDNA.16089.1.S1_at | Glutamate Receptor, Ionotropic, (KA1)/(Kainate 4) | Grik4 | − | − |
| MmugDNA.3852.1.S1_at | Glutamate Receptor (gamma2), Ionotropic (KA2)/(Kainate 5) | Grik5 | − | + |
| MmugDNA.8027.1.S1_at | Glutamate Receptor (gamma2), Ionotropic (KA2)/(Kainate 5) | Grik5 | − | − |
| MmuSTS.3653.1.S1_at | Glutamate Receptor, Ionotropic (GluRdelta1)/(delta1) | Grid1 | − | − |
| MmuSTS.3678.1.S1_at | Glutamate Receptor, Ionotropic (GluRdelta2)/(delta2) | Grid2 | − | − |
Table legend: “+” and “−” symbols represent values obtained using Affymetrix GCOS analysis, and global scaling to an average target intensity of 200. (−) = “undetectable”, (+/−) = “marginally present”, (+) = “signal intensity < 500”, (++) = “signal intensity 500–1000”, (+++) = “signal intensity > 1000”.
Table 3.
Expression of metabotropic glutamate receptor subunit mRNA in the rhesus macaque locus coeruleus
| Affymetrix Probe Set ID | Common Gene Description | Gene Symbol | Animal 1 | Animal 2 |
|---|---|---|---|---|
| MmugDNA.22055.1.S1_at | Glutamate Receptor, Metabotropic 1, (mGluR1) | Grm1 | + | ++ |
| MmugDNA.32554.1.S1_at | Glutamate Receptor, Metabotropic 1, (mGluR1) | Grm1 | − | − |
| MmugDNA.16347.1.S1_at | Glutamate Receptor, Metabotropic 2, (mGluR2) | Grm2 | − | − |
| MmugDNA.37104.1.S1_at | Glutamate Receptor, Metabotropic 2, (mGluR2) | Grm2 | − | − |
| MmugDNA.16349.1.S1_at | Glutamate Receptor, Metabotropic 3, (mGluR3) | Grm3 | +++ | +++ |
| MmugDNA.26361.1.S1_at | Glutamate Receptor, Metabotropic 3, (mGluR3) | Grm3 | − | − |
| MmugDNA.32363.1.S1_at | Glutamate Receptor, Metabotropic 3, (mGluR3) | Grm3 | − | − |
| MmuSTS.4.1.S1_at | Glutamate Receptor, Metabotropic 4, (mGluR4) | Grm4 | + | + |
| MmugDNA.16371.1.S1_at | Glutamate Receptor, Metabotropic 5, (mGluR5) | Grm5 | + | − |
| MmugDNA.22462.1.S1_at | Glutamate Receptor, Metabotropic 5, (mGluR5) | Grm5 | − | − |
| MmugDNA.9291.1.S1_at | Glutamate Receptor, Metabotropic 5, (mGluR5) | Grm5 | ++ | ++ |
| MmugDNA.22057.1.S1_at | Glutamate Receptor, Metabotropic 6, (mGluR6) | Grm6 | − | + |
| MmugDNA.10814.1.S1_at | Glutamate Receptor, Metabotropic 7, (mGluR7) | Grm7 | − | +/− |
| MmugDNA.14760.1.S1_at | Glutamate Receptor, Metabotropic 7, (mGluR7) | Grm7 | − | − |
| MmugDNA.31046.1.S1_at | Glutamate Receptor, Metabotropic 7, (mGluR7) | Grm7 | + | + |
| MmugDNA.11213.1.S1_at | Glutamate Receptor, Metabotropic 8, (mGluR8) | Grm8 | − | − |
| MmugDNA.20667.1.S1_at | Glutamate Receptor, Metabotropic 8, (mGluR8) | Grm8 | − | − |
Table legend: “+” and “−” symbols represent values obtained using Affymetrix GCOS analysis, and global scaling to an average target intensity of 200. (−) = “undetectable”, (+/−) = “marginally present”, (+) = “signal intensity < 500”, (++) = “signal intensity 500–1000”, (+++) = “signal intensity > 1000”.
3. Discussion
The presence of mRNA encoding tyrosine hydroxylase (TH), a rate-limiting enzyme of the catecholamine biosynthesis pathway, indicates that the micro-dissected tissues used in the micorarray analysis were catecholaminergic, while the complete absence of mRNA encoding GFAP suggests a negligible level of contamination of the extracted LC RNA with mRNA of glial origin. Exactly why Animals 1 and 2 showed different levels of TH expression is unclear, but may stem from differences in physiological states of the two individuals. NE levels in the LC show circadian variation (Agren et al., 1986) and TH mRNA has been shown to increase in the LC of rats subjected to REM sleep deprivation (Porkka-Heiskanen et al., 1995). It is possible that the level of LC activity and NE release may have differed between the two monkeys according to their activity pattern, waking state and level of stimulation (Cirelli et al., 1996; Aston-Jones et al., 2000; Berridge and Waterhouse, 2003). On the other hand, closely matching GAPDH and ACTB expression levels in both animals imply that the expression levels observed for other genes were comparable between the two animals.
Our microarray data demonstrates that the rhesus macaque LC expresses genes that encode VGLUTs, which play a key role in glutamatergic neurotransmission by loading glutamate into presynaptic vesicles (Bellocchio et al., 1998; Bellocchio et al., 2000; Takamori et al., 2000). VGLUT1 not only confers glutamate uptake activity to synaptic vesicles but is also considered sufficient to define a glutamatergic phenotype in neurons (Takamori, 2006). Noxious, stressful corticoid and cardiovascular stimulation as well as opiate withdrawal resulting in glutamate efflux localized in the LC has been described using microdialysis and/or push-pull superfusion studies in anaesthetized rats (Aghajanian et al., 1994; Singewald et al., 1994), conscious rats (Zhang et al., 1994; Feng et al., 1995; Hoshi et al., 1996; Tokuyama and Ho, 1996; Feng et al., 1997; Hoshi et al., 1997) and the conscious cat (Nitz and Siegel, 1997). Although the efflux of glutamate from the LC has been attributed to release from afferents, the expression of glutaminase and VGLUT mRNA in the current study suggests that glutamate may also be synthesized and released within the LC itself. Similarly, the high expression of glutamic acid decarboxylase (GAD) mRNA in the LC suggests that the some of the endogenous glutamate may ultimately be converted into GABA.
The microarray analysis corroborated the immunohistochemical and in situ hybridization findings by showing that NR1, GluR1, GluR2, and GluR3 subunit mRNA is expressed in the rhesus macaque LC. The housekeeping genes GAPDH and ACTB are described as having a “moderately abundant” message (Bustin, 2000). From this reference description, GluR3 expression could also be called moderately abundant, whereas gene expression of NR1 GluR1 and GluR2 could be described as “low to moderate”. However such expression differences were not evident from visual inspection of in situ images where expression was clear and at roughly equivalent intensity between the subunits. Interestingly, some of the microarray’s NR1 probe sets failed to disclose significant levels of NR1 gene expression. Although the exact reason for this is unclear, it could reflect the existence of multiple NR1 isoforms in the monkey LC. In rats, the NR1 subunit has eight isoforms, which are generated from alternative splicing of the NR1 gene sequence (Zukin and Bennett, 1995). If similar splice variation occurs in the rhesus macaque LC it would reduce the hybridization probability of any given probe set. In addition, if expression of specific NR1 splice variants is limited, it could explain why only one of three probe sets showed significant hybridization using the rhesus macaque microarray.
The microarray analysis also showed expression of NR2A and NR2D receptor subunits in the LC. Although NR1 is considered to be the obligatory subunit of the multimeric NMDA receptor, overall function of this receptor is also dependent upon the presence of NR2 subunits (Ishii et al., 1993; Monyer et al., 1994; Behe et al., 1995). Taken together, therefore, the presence of NR1, NR2A and NR2D supports the view that functional NMDA receptors are present within the rhesus macaque LC. Western blot analyses of LC homogenate from a human subject showed high immunoreactivity to NR1 and NR2C subunits and low immunoreactivity to NR2A and NR2B, whereas NR2C levels appeared elevated among depressive subjects (Karolewicz et al., 2005). More recent evidence indicates that NR1 subunit expression is reduced in the LC of postmortem alcoholic human subjects compared to controls (Karolewicz et al., 2007). In the current study, NR2C subunit mRNA expression appeared possible in one of the two animals, but NR2B mRNA expression could not be detected. Together these data suggest that functional ionotropic NMDA glutamate receptors are present in the primate LC with the possibility of variability in subunit composition. Similarly, our disclosure of mRNA encoding all four AMPA receptor subunits (i.e., GluR1, GluR2, GluR3, and GluR4) suggests that functional AMPA receptors are also present.
In contrast, mRNA expression of the subunit components necessary for functional kainate receptors appears to be limited in the rhesus macaque LC. The only kainate receptor subunit with significant RNA expression was GluR6, while concomitant expression of other kainate receptor subunits (i.e., GluR5, GluR7, KA1 and KA2) appeared to be absent. Although homomeric GluR6 subunit expression is sufficient for the formation of a functional low-affinity kainate receptor (Sommer and Seeburg, 1992), heteromeric expression with other subunits (KA1 and KA2) is necessary for the formation of high-affinity kainite receptors.
Confirmation of NR1 expression in the rhesus macaque LC by in situ hybridization and immunohistochemistry, in this and previous studies (Luque et al., 1995; Urbanski et al., 1997), as well as the current demonstration of both NR1 and NR2 gene expression by gene microarray analysis, are consistent with the reported localization of NR1 and NR2 receptor subunits in the rat LC (Luque et al., 1995). The NR1 subunit is widely expressed in the rat brain (Moriyoshi et al., 1991; Monyer et al., 1992) as well as in the rhesus macaque (Garyfallou et al., 1996; Kohama and Urbanski, 1997; Kohama et al., 1998) and human brain (Rigby et al., 1996). Other NMDA subunits are more regionally restricted and specific combinations of NR1 (obligatory) subunits with NR2 and/or NR3 subunits determine the functional properties of a given NMDA receptor (Meguro et al., 1992; Ishii et al., 1993; Wafford et al., 1993; Sucher et al., 1995; Das et al., 1998; Vallano, 1998; Goebel and Poosch, 1999). Because we focused our study on adult animals only we cannot exclude the interesting possibility that changes in NMDA receptors may vary with developmental stimulus, as has been described in the visual cortex (Fox et al., 1992), and thereby modulate LC sensitivity to glutamatergic inputs.
Similarly, the relative gene expression levels that we observed for AMPA receptor subunits are consistent with previous in situ hybridization data from the rat (Tohyama and Oyamada, 1994), where mRNAs encoding GluR1, GluR2, GluR3, and GluR4 AMPA receptor subunits were all expressed in the LC, and GluR2 mRNA expression was found to be the strongest. Although the exact functional role of AMPA receptors in the LC is unclear, the use of glutamate receptor antagonists to reduce LC activation resulting from opiate withdrawal indicates that AMPA receptors play a more active role than kainate or NMDA receptors (Rasmussen, 1995; Rasmussen et al., 1996).
Interestingly, the LC expressed mRNA sequences for five of the eight metabotropic glutamate receptor subunits, suggesting that glutamatergic influence within this nucleus is likely to be mediated not only by NMDA and AMPA ionotropic receptors but also by the metabotropic receptors. Of the known metabotropic receptors (which make use of second messenger systems), the mGluR5 receptor appears to play a role in activating the rat LC after opiate withdrawal (Rasmussen et al., 2005), which is consistent with our observation that mGluR5 mRNA is highly expressed in the rhesus macaque LC. In addition, the mGluR2/3 receptor antagonist LY341495 reduces LC activity in response to opiate-withdrawal-induced activation (Vandergriff and Rasmussen, 1999; Rasmussen et al., 2004), which is also consistent with our observation that mGluR3 mRNA, but not mGluR2 mRNA, is also highly expressed in the primate LC. However it should be noted that members of the mGluR family are further classified into groups according to sequence similarities, signal transduction mechanisms and agonist selectivities. Currently mGluR 2 and 3 belong to the same mGluR2/3 subgroup, which has very different agonist selectivity compared to the mGluR1/5 or the mGluR4/6/7 subgroups (Nakanishi, 1992; Nakanishi and Masu, 1994; Nakanishi et al., 1994). Under this classification system, a representative from each of the three mGluR subgroups was expressed in the rhesus LC based upon microarray analysis. However, more selective agonists are required for more detailed study of the mGluR subunits.
As emphasized above, the present study represents a first attempt at comprehensive characterization of glutamate receptor expression in the LC of a primate species. The results should have clinical relevance, given that changes in glutamate receptor subunit composition play a role in synaptic plasticity (Olson and Freeman, 1980; Vallano, 1998; Erreger et al., 2005; Kohr, 2006; Bartlett et al., 2007)), and are targets of pharmacological research aimed at treating Alzheimer’s disease, schizophrenia and depression (O’Neill et al., 2004; Black, 2005; Palucha and Pilc, 2005; Planells-Cases et al., 2006; Beneyto et al., 2007).
4. Experimental Procedures
4.1. Animals
Adult rhesus macaques (Macaca mulatta) were cared for in accordance with the NIH Guide for the Care and Use of Laboratory Animals by the Division of Animal Resources at the Oregon National Primate Research Center (ONPRC). Ultimately they were euthanized with sodium pentobarbital and provided postmortem tissue for this and other studies, via the ONPRC Tissue Distribution Program (TDP). To obtain hindbrain tissue suitable for immunohistochemistry and in situ hybridization, the brains were perfused via aortic cannulation with 1 liter of 0.9% saline followed by 6.5 liters of ice-cold 4% paraformaldehyde in 3.8% sodium tetraborate buffer (pH 9.5). For GeneChip® microarray profiling, brains from two additional ovariectomized adult rhesus macaques were obtained through the TDP. After perfusion via with 1 liter of 0.9% saline, these brains were immediately removed and blocked at the rostral juncture of the cerebellum and midbrain. The tissue was preserved in RNAlater® (Ambion; Austin, TX) and the LC was subsequently microdissected from the surrounding tissue.
4.2. Tissue preparation for immunohistochemistry and in situ hybridization
For immunohistochemistry and in situ hybridization, the brains were removed immediately after paraformaldehyde perfusion and the hindbrain was immersed in fresh fixative for an additional 3 h at 4°C. The tissue was cryoprotected, first by immersion for 24 h in a 10% glycerol solution in 0.02 M phosphate buffer containing 2% dimethyl sulfoxide (DMSO) and then by immersion for an additional 72 h in a similar phosphate/DMSO solution containing 20% glycerol. The tissue was then rapidly frozen in 2-methylbutane that was pre-cooled in an ethanol/dry-ice bath, and stored at −85°C. Subsequently, frozen coronal sections (25 μm) were cut using a sliding microtome and were processed either for immunohistochemistry or in situ hybridization. In the latter case, the sections were mounted on glass microscope slides (Fisherbrand Superfrost/Plus; Fisher, Auburn, WA), air dried for 30 min, vacuum dried overnight, and then stored at −85°C for later use.
4.3 Immunohistochemistry
To identify the location of the LC, immunohistochemistry was performed on selected free-floating brain sections using a mouse monoclonal antibody specific to tyrosine hydroxylase (TH) (Boehringer-Mannheim Corp., Indianapolis, IN), as previously demonstrated (Urbanski et al., 1997). To reduce the level of non-specific immuno-labeling the sections were pre-incubated for at least 1 h at room temperature in a 50 mM Tris buffer (pH 7.6) containing 0.9% NaCl, 0.1% Triton® X-100 (Tris A), and 2% normal horse serum. They were then incubated overnight at 4°C with the TH antibody at a dilution of 1:500 in Tris A. Sections adjacent to TH-containing sections were immunostained for NMDA and AMPA ionotropic glutamate receptor subunits, using rabbit polyclonal antibodies against NR1, GluR1, and GluR2/3 (Chemicon Int; Temecula, CA). Each of these antibodies has been shown to be immunospecific to the C-terminus of the corresponding rat receptor subunits, and the NR1 antiserum is thought to recognize four of the seven splice variants (i.e., R1A, R1B, R1C, AND R1F) of the rat NR1 receptor subunit (Petralia and Wenthold, 1992; Wenthold et al., 1992; Petralia et al., 1994). Each of these polyclonal antibodies has been previously validated for use with rhesus macaque brain tissue (Garyfallou et al., 1996) (Kohama and Urbanski, 1997). They were used at a dilution of 1–4 μg/ml in Tris A and the primary incubations were performed at 4°C for 30 h on an orbital shaker. Sections were then washed Tris A (3×10 min) and incubated for 1 h at room temperature with a biotinylated antibody. Biotinylated horse anti-mouse IgG (1:1000 dilution, Vector Laboratories, Burlingame, CA) was used for TH staining, and biotinylated goat anti-rabbit IgG (1:1000 dilution, Vector Laboratories) was used for glutamate receptor subunit staining. Sections were then washed again in Tris A (3×10 min) and incubated for 1.5 h with a peroxidase-coupled avidin-biotin complex (Peroxidase-ABC Kit; Vector Laboratories). After further washing with Tris A, the sections were incubated in Tris-saline solution containing 3,3′-diaminobenzidine tetrahydrochloride (DAB;10 mg/ml) and 0.003% H2O2 for approximately 10 min. They were then rinsed with Tris-saline buffer, mounted on glass microscope slides (Fisherbrand Superfrost/Plus), air-dried, dehydrated, cleared, and coverslipped using DPX mounting medium (Electron Microscopy Sciences, Ft. Washington, PA).
4.4 In situ hybridization
Although the GluR2/3 antibody used for the immunohistochemical study produced dense immunostaining, it was unclear from these observations if the Glu2, Glu3 or both of the two AMPA receptor subunits was being expressed. In order to distinguish between the GluR2 and GluR3 receptor subunits and to corroborate the other immunohistochemical findings, in situ hybridization was performed on sections adjacent to those showing immuno-positive labeling of TH. Specific anti-sense 35S-labeled cRNA probes were transcribed from rat cDNAs encoding the NR1, GluR1, GluR2 and GluR3 glutamate receptor subunits. The lengths of the riboprobes varied from 300–600 nucleotides. The NR1 antisense probe (450 nucleotides) corresponded to the region between the PstI and EcoRI restriction sites of the NR1 cDNA (Moriyoshi et al., 1991). The GluR1, GluR2 and GluR3 antisense probes were transcribed from the corresponding Flop cDNAs, which were cut at the XmnI, SphI and SalI restriction sites respectively (i.e., 273, 428 and 623 nucleotides from the 3′ end of the corresponding cDNA) (Hollmann et al., 1989; Boulter et al., 1990). In each case, synthesis of the RNA transcripts was performed following the instructions supplied with a transcription kit (Promega, Madison, WI). Each of these glutamate receptor subunit riboprobes has previously been validated for use in paraformaldehyde-fixed rhesus macaque brain tissue (Garyfallou et al., 1996; Kohama et al., 1998).
The hindbrain sections were post-fixed in 4% paraformaldehyde, 0.1 M phosphate buffer, pH 7.4, for 15 min, rinsed in Tris-EDTA, and then were treated with proteinase K (10 μg/ml) in Tris-EDTA buffer (pH 8.0) for 30 min. Next, they were acetylated, dehydrated with ethanol, dried under vacuum for 2 h, and then hybridized overnight at 60°C with 25 μl of antisense 35S-labeled riboprobe diluted to 10×106 c.p.m. per ml of hybridization buffer (50 mM dithiothreitol, 250 μg/ml tRNA, 50% formamide, 0.3 M NaCl, 1× Denhardt’s solution, 20 mM Tris (pH 8.0), 1 mM EDTA (pH 8.0), and 10% dextran sulfate). The post-hybridization step involved washing off coverslips in 4x saline-sodium citrate buffer (SSC; the 20× stock solution comprised 175.3 g sodium chloride and 88.2 g sodium citrate per liter (pH 7.0)) containing 20 mM dithiothreitol. The sections were then incubated in Tris-EDTA buffer (pH 8.0) containing RNase A (10 μg/ml) for 30 min at 37°C, followed by two 30-min washes with 2× SSC at room temperature, and a final wash with 0.1× SSC at 70°C. The mounted sections were then dehydrated using an ascending ethanol gradient, containing 0.3 M ammonium acetate, and air-dried for 30 min. To visualize the hybridization pattern of the 35S-labeled RNA probe, the section were dipped in NTB-2 photographic emulsion (Eastman Kodak Company, Rochester, NY) and exposed for five days at 4°C in a light-tight box. The mounted sections were subsequently processed with Kodak developer (D-19) and fixer, counterstained with 0.1% thionin, dehydrated with ethanol, cleared with xylenes, and finally coverslipped using DPX mounting medium (Electron Microscopy Sciences).
4.5 Hybridization to Affymetrix Rhesus Genome GeneChip® Array
In order to further examine expression of glutamate receptor subunit genes in the LC, we conducted a microarray analysis of rhesus macaque LC. Whole RNA was extracted from the tissues using the Qiagen RNeasy mini kit, following the manufacturer’s instructions. Briefly, tissues were homogenized in RLT buffer with 1% β-mercaptoethanol. RNA was collected onto RNeasy mini columns and eluted in water, where quality was assessed using an Agilent 2100 Bioanalyzer. For both samples, the RNA quality exceeded the level recommended by Affymetrix for hybridization to the Rhesus Genome GeneChip® (micro)array.
Microarray assays were performed in the Affymetrix Microarray Core (AMC) of the OHSU Gene Microarray Shared Resource. Samples were prepared using the AMC One-cycle cDNA IVT (in vitro transcription) amplification/labeling protocol (Standard Labeling). Each sample target was hybridized to a Rhesus Genome GeneChip® microarray and image processing and expression analysis were performed using Affymetrix GeneChip® Operating Software (GCOS) version 1.2. After GCOS analysis, absolute analyses were re-run using global scaling to an average target intensity of 200. In this way, scaling allowed for direct comparison of the hybridization values from the two targets. The parameters α1 and α2, which set the point at which the probe set is called present, marginally present, or undetectable, were set to 0.05 and 0.065 (Affymetrix defaults) respectively. These parameters were used to determine expression levels for each probe set based on the detection P-value of the probe set. An assay performance assessment was conducted to confirm that the quality of the RNA used was acceptable and that genome hybridizations performed well. The performance assessment (conducted by AMC) was based on the GCOS analysis, and pair-wise scatter plots of Affymetrix Microarray Suite (MAS) absolute analyses. We used Affymetrix probe set ID numbers to search the GeneChip® microarray for oligonucleotide signal intensities associated with genes for subunits of ionotropic glutamate receptor genes from the NMDA, AMPA and kainate families as well as orphan (delta) subunits (Dingledine et al., 1999). Probe set ID assignments were referenced according to listings for the Rhesus GeneChip® microarray and searched using the Affymetrix online Netaffx™ Analysis Center software (query function). We considered a probe set to be expressed if it was detected as “present” in both animals.
In addition to searching the GeneChip® database for glutamate receptor subunits we used these methods to confirm the specificity of our LC mRNA by examining gene expression for tyrosine hydroxylase (TH), which is indicative of catecholamine synthesis and so reliably identifies cells within the LC but not in the surrounding pericoerulear tissue. The pericoerulear tissue has a high concentration of glial cells relative to the LC; therefore as a negative control, we examined GFAP expression to test for RNA contamination from cells surrounding the LC (Table 1). We also focused on the expression of genes encoding VGLUT and glutaminase (to explore the hypothesis that the neurotransmitter glutamate is itself synthesized within the LC), and on glutamic acid decarboxylase (GAD) (to explore the possibility that endogenous glutamate is converted to GABA within the LC). Lastly, we examined the expression of ACTB and GAPDH as a signal intensity reference because these housekeeping genes are reliably expressed in many tissues under various conditions, and are used as standards for normalizing errors in mRNA quantification (Bustin, 2000; Suzuki et al., 2000).
Acknowledgments
This work was supported by NIH grants: HD-29186 and RR-00163. Gene microarray assays were performed in the Affymetrix Microarray Core of the Gene Microarray Shared Resource facility at OHSU.
Abbreviations
- ACTB
β-actin
- AMC
Affymetrix Microarray Core
- AMPA
α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid
- DAB
3,3′-diaminobenzidine tetrahydrochloride
- DMSO
dimethyl sulfoxide
- E
estrogen
- EAA
excitatory amino acid
- GABA
γ-amino butyric acid
- GAD
glutamic acid decarboxylase
- GAPDH
glyceraldehyde 3-phosphate dehydrogenase
- GCOS
GeneChip® Operating Software
- GFAP
glial fibrillary acidic protein
- GnRH
gonadotropin releasing hormone
- LC
locus coeruleus
- NE
norepinephrine
- NET
norepinephrine transporter
- NMDA
N-methyl-D-aspartate
- OHSU
Oregon Health & Science University
- ONPRC
Oregon National Primate Research Center
- TH
tyrosine hydroxylase
- TDP
Tissue Distribution Program
- VGLUT
vesicular glutamate transporter
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Literature references
- Abercrombie ED, Jacobs BL. Single-unit response of noradrenergic neurons in the locus coeruleus of freely moving cats. I Acutely presented stressful and nonstressful stimuli. J Neurosci. 1987;7:2837–2843. doi: 10.1523/JNEUROSCI.07-09-02837.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aghajanian GK, Kogan JH, Moghaddam B. Opiate withdrawal increases glutamate and aspartate efflux in the locus coeruleus: an in vivo microdialysis study. Brain Res. 1994;636:126–130. doi: 10.1016/0006-8993(94)90186-4. [DOI] [PubMed] [Google Scholar]
- Agren H, Koulu M, Saavedra JM, Potter WZ, Linnoila M. Circadian covariation of norepinephrine and serotonin in the locus coeruleus and dorsal raphe nucleus in the rat. Brain Res. 1986;397:353–358. doi: 10.1016/0006-8993(86)90638-4. [DOI] [PubMed] [Google Scholar]
- Aston-Jones G, Bloom FE. Nonrepinephrine-containing locus coeruleus neurons in behaving rats exhibit pronounced responses to non-noxious environmental stimuli. J Neurosci. 1981;1:887–900. doi: 10.1523/JNEUROSCI.01-08-00887.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aston-Jones G, Shipley MT, Chouvet G, Ennis M, van Bockstaele E, Pieribone V, Shiekhattar R, Akaoka H, Drolet G, Astier B, et al. Afferent regulation of locus coeruleus neurons: anatomy, physiology and pharmacology. Prog Brain Res. 1991;88:47–75. doi: 10.1016/s0079-6123(08)63799-1. [DOI] [PubMed] [Google Scholar]
- Aston-Jones G, Rajkowski J, Cohen J. Locus coeruleus and regulation of behavioral flexibility and attention. Progress in brain research. 2000;126:165–182. doi: 10.1016/S0079-6123(00)26013-5. [DOI] [PubMed] [Google Scholar]
- Aston-Jones G. Brain structures and receptors involved in alertness. Sleep medicine. 2005;6(Suppl 1):S3–7. doi: 10.1016/s1389-9457(05)80002-4. [DOI] [PubMed] [Google Scholar]
- Aston-Jones G, Cohen JD. An integrative theory of locus coeruleus-norepinephrine function: adaptive gain and optimal performance. Annu Rev Neurosci. 2005a;28:403–450. doi: 10.1146/annurev.neuro.28.061604.135709. [DOI] [PubMed] [Google Scholar]
- Aston-Jones G, Cohen JD. Adaptive gain and the role of the locus coeruleus-norepinephrine system in optimal performance. J Comp Neurol. 2005b;493:99–110. doi: 10.1002/cne.20723. [DOI] [PubMed] [Google Scholar]
- Bartlett TE, Bannister NJ, Collett VJ, Dargan SL, Massey PV, Bortolotto ZA, Fitzjohn SM, Bashir ZI, Collingridge GL, Lodge D. Differential roles of NR2A and NR2B-containing NMDA receptors in LTP and LTD in the CA1 region of two-week old rat hippocampus. Neuropharmacology. 2007;52:60–70. doi: 10.1016/j.neuropharm.2006.07.013. [DOI] [PubMed] [Google Scholar]
- Behe P, Stern P, Wyllie DJ, Nassar M, Schoepfer R, Colquhoun D. Determination of NMDA NR1 subunit copy number in recombinant NMDA receptors. Proc Biol Sci. 1995;262:205–213. doi: 10.1098/rspb.1995.0197. [DOI] [PubMed] [Google Scholar]
- Bellocchio EE, Hu H, Pohorille A, Chan J, Pickel VM, Edwards RH. The localization of the brain-specific inorganic phosphate transporter suggests a specific presynaptic role in glutamatergic transmission. J Neurosci. 1998;18:8648–8659. doi: 10.1523/JNEUROSCI.18-21-08648.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellocchio EE, Reimer RJ, Fremeau RT, Jr, Edwards RH. Uptake of glutamate into synaptic vesicles by an inorganic phosphate transporter. Science. 2000;289:957–960. doi: 10.1126/science.289.5481.957. [DOI] [PubMed] [Google Scholar]
- Beneyto M, Kristiansen LV, Oni-Orisan A, McCullumsmith RE, Meador-Woodruff JH. Abnormal Glutamate Receptor Expression in the Medial Temporal Lobe in Schizophrenia and Mood Disorders. Neuropsychopharmacology. 2007 doi: 10.1038/sj.npp.1301312. Epub ahead of print. http://www.nature.com/npp/journal/vaop/ncurrent/pdf/1301312a.pdf. [DOI] [PubMed]
- Berridge CW, Waterhouse BD. The locus coeruleus-noradrenergic system: modulation of behavioral state and state-dependent cognitive processes. Brain Res Brain Res Rev. 2003;42:33–84. doi: 10.1016/s0165-0173(03)00143-7. [DOI] [PubMed] [Google Scholar]
- Black MD. Therapeutic potential of positive AMPA modulators and their relationship to AMPA receptor subunits. A review of preclinical data. Psychopharmacology (Berl) 2005;179:154–163. doi: 10.1007/s00213-004-2065-6. [DOI] [PubMed] [Google Scholar]
- Boulter J, Hollmann M, O’Shea-Greenfield A, Hartley M, Deneris E, Maron C, Heinemann S. Molecular cloning and functional expression of glutamate receptor subunit genes. Science. 1990;249:1033–1037. doi: 10.1126/science.2168579. [DOI] [PubMed] [Google Scholar]
- Bourgin P, Huitron-Resendiz S, Spier AD, Fabre V, Morte B, Criado JR, Sutcliffe JG, Henriksen SJ, de Lecea L. Hypocretin-1 modulates rapid eye movement sleep through activation of locus coeruleus neurons. Journal of neuroscience. 2000;20:7760–7765. doi: 10.1523/JNEUROSCI.20-20-07760.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brann DW. Glutamate: a major excitatory transmitter in neuroendocrine regulation. Neuroendocrinology. 1995;61:213–225. doi: 10.1159/000126843. [DOI] [PubMed] [Google Scholar]
- Brann DW, Mahesh VB. Glutamate: a major neuroendocrine excitatory signal mediating steroid effects on gonadotropin secretion. Journal of steroid biochemistry and molecular biology. 1995;53:325–329. doi: 10.1016/0960-0760(95)00070-g. [DOI] [PubMed] [Google Scholar]
- Bustin SA. Absolute quantification of mRNA using real-time reverse transcription polymerase chain reaction assays. Journal of molecular endocrinology. 2000;25:169–193. doi: 10.1677/jme.0.0250169. [DOI] [PubMed] [Google Scholar]
- Cedarbaum JM, Aghajanian GK. Activation of locus coeruleus neurons by peripheral stimuli: modulation by a collateral inhibitory mechanism. Life Sci. 1978;23:1383–1392. doi: 10.1016/0024-3205(78)90398-3. [DOI] [PubMed] [Google Scholar]
- Cirelli C, Pompeiano M, Tononi G. Neuronal gene expression in the waking state: a role for the locus coeruleus. Science. 1996;274:1211–1215. doi: 10.1126/science.274.5290.1211. [DOI] [PubMed] [Google Scholar]
- Clayton EC, Rajkowski J, Cohen JD, Aston-Jones G. Phasic activation of monkey locus ceruleus neurons by simple decisions in a forced-choice task. J Neurosci. 2004;24:9914–9920. doi: 10.1523/JNEUROSCI.2446-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunningham ET, Jr, Sawchenko PE. Anatomical specificity of noradrenergic inputs to the paraventricular and supraoptic nuclei of the rat hypothalamus. Journal of comparative neurology. 1988;274:60–76. doi: 10.1002/cne.902740107. [DOI] [PubMed] [Google Scholar]
- Curtis AL, Drolet G, Valentino RJ. Hemodynamic stress activates locus coeruleus neurons of unanesthetized rats. Brain Res Bull. 1993;31:737–744. doi: 10.1016/0361-9230(93)90150-a. [DOI] [PubMed] [Google Scholar]
- Das S, Sasaki YF, Rothe T, Premkumar LS, Takasu M, Crandall JE, Dikkes P, Conner DA, Rayudu PV, Cheung W, Chen HS, Lipton SA, Nakanishi N. Increased NMDA current and spine density in mice lacking the NMDA receptor subunit NR3A. Nature. 1998;393:377–381. doi: 10.1038/30748. [DOI] [PubMed] [Google Scholar]
- Dingledine R, Borges K, Bowie D, Traynelis SF. The glutamate receptor ion channels. Pharmacol Rev. 1999;51:7–61. [PubMed] [Google Scholar]
- Downs JL, Dunn MR, Borok E, Shanabrough M, Horvath TL, Kohama SG, Urbanski HF. Orexin neuronal changes in the locus coeruleus of the aging rhesus macaque. Neurobiol Aging. 2007;28:1286–1295. doi: 10.1016/j.neurobiolaging.2006.05.025. [DOI] [PubMed] [Google Scholar]
- Elam M, Svensson TH, Thoren P. Differentiated cardiovascular afferent regulation of locus coeruleus neurons and sympathetic nerves. Brain Res. 1985;358:77–84. doi: 10.1016/0006-8993(85)90950-3. [DOI] [PubMed] [Google Scholar]
- Erreger K, Dravid SM, Banke TG, Wyllie DJ, Traynelis SF. Subunit-specific gating controls rat NR1/NR2A and NR1/NR2B NMDA channel kinetics and synaptic signalling profiles. J Physiol. 2005;563:345–358. doi: 10.1113/jphysiol.2004.080028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espana RA, Reis KM, Valentino RJ, Berridge CW. Organization of hypocretin/orexin efferents to locus coeruleus and basal forebrain arousal-related structures. Journal of comparative neurology. 2005;481:160–178. doi: 10.1002/cne.20369. [DOI] [PubMed] [Google Scholar]
- Feng Y, Rockhold RW, Ho IK. Nor-binaltorphimine precipitates withdrawal and excitatory amino acid release in the locus ceruleus of butorphanol--but not morphine-dependent rats. J Pharmacol Exp Ther. 1997;283:932–938. [PubMed] [Google Scholar]
- Feng YZ, Zhang T, Rockhold RW, Ho IK. Increased locus coeruleus glutamate levels are associated with naloxone-precipitated withdrawal from butorphanol in the rat. Neurochem Res. 1995;20:745–751. doi: 10.1007/BF01705544. [DOI] [PubMed] [Google Scholar]
- Foote SL, Bloom FE, Aston-Jones G. Nucleus locus ceruleus: new evidence of anatomical and physiological specificity. Physiol Rev. 1983;63:844–914. doi: 10.1152/physrev.1983.63.3.844. [DOI] [PubMed] [Google Scholar]
- Fruhstorfer B, Mignot E, Bowersox S, Nishino S, Dement WC, Guilleminault C. Canine narcolepsy is associated with an elevated number of alpha 2-receptors in the locus coeruleus. Brain research. 1989;500:209–214. doi: 10.1016/0006-8993(89)90315-6. [DOI] [PubMed] [Google Scholar]
- Garyfallou VT, Kohama SG, Urbanski HF. Distribution of NMDA and AMPA receptors in the cerebellar cortex of rhesus macaques. Brain Res. 1996;716:22–28. doi: 10.1016/0006-8993(95)01545-0. [DOI] [PubMed] [Google Scholar]
- Gasic GP, Hollmann M. Molecular neurobiology of glutamate receptors. Annu Rev Physiol. 1992;54:507–536. doi: 10.1146/annurev.ph.54.030192.002451. [DOI] [PubMed] [Google Scholar]
- Gay VL, Plant TM. Sustained intermittent release of gonadotropin-releasing hormone in the prepubertal male rhesus monkey induced by N-methyl-DL-aspartic acid. Neuroendocrinology. 1988;48:147–152. doi: 10.1159/000125002. [DOI] [PubMed] [Google Scholar]
- Gearing M, Terasawa E. The alpha-1-adrenergic neuronal system is involved in the pulsatile release of luteinizing hormone-releasing hormone in the ovariectomized female rhesus monkey. Neuroendocrinology. 1991;53:373–381. doi: 10.1159/000125744. [DOI] [PubMed] [Google Scholar]
- Gibbs RA, Rogers J, Katze MG, Bumgarner R, Weinstock GM, Mardis ER, Remington KA, Strausberg RL, Venter JC, Wilson RK, Batzer MA, Bustamante CD, Eichler EE, Hahn MW, Hardison RC, Makova KD, Miller W, Milosavljevic A, Palermo RE, Siepel A, Sikela JM, Attaway T, Bell S, Bernard KE, Buhay CJ, Chandrabose MN, Dao M, Davis C, Delehaunty KD, Ding Y, Dinh HH, Dugan-Rocha S, Fulton LA, Gabisi RA, Garner TT, Godfrey J, Hawes AC, Hernandez J, Hines S, Holder M, Hume J, Jhangiani SN, Joshi V, Khan ZM, Kirkness EF, Cree A, Fowler RG, Lee S, Lewis LR, Li Z, Liu YS, Moore SM, Muzny D, Nazareth LV, Ngo DN, Okwuonu GO, Pai G, Parker D, Paul HA, Pfannkoch C, Pohl CS, Rogers YH, Ruiz SJ, Sabo A, Santibanez J, Schneider BW, Smith SM, Sodergren E, Svatek AF, Utterback TR, Vattathil S, Warren W, White CS, Chinwalla AT, Feng Y, Halpern AL, Hillier LW, Huang X, Minx P, Nelson JO, Pepin KH, Qin X, Sutton GG, Venter E, Walenz BP, Wallis JW, Worley KC, Yang SP, Jones SM, Marra MA, Rocchi M, Schein JE, Baertsch R, Clarke L, Csuros M, Glasscock J, Harris RA, Havlak P, Jackson AR, Jiang H, Liu Y, Messina DN, Shen Y, Song HX, Wylie T, Zhang L, Birney E, Han K, Konkel MK, Lee J, Smit AF, Ullmer B, Wang H, Xing J, Burhans R, Cheng Z, Karro JE, Ma J, Raney B, She X, Cox MJ, Demuth JP, Dumas LJ, Han SG, Hopkins J, Karimpour-Fard A, Kim YH, Pollack JR, Vinar T, Addo-Quaye C, Degenhardt J, Denby A, Hubisz MJ, Indap A, Kosiol C, Lahn BT, Lawson HA, Marklein A, Nielsen R, Vallender EJ, Clark AG, Ferguson B, Hernandez RD, Hirani K, Kehrer-Sawatzki H, Kolb J, Patil S, Pu LL, Ren Y, Smith DG, Wheeler DA, Schenck I, Ball EV, Chen R, Cooper DN, Giardine B, Hsu F, Kent WJ, Lesk A, Nelson DL, O’Brien WE, Prufer K, Stenson PD, Wallace JC, Ke H, Liu XM, Wang P, Xiang AP, Yang F, Barber GP, Haussler D, Karolchik D, Kern AD, Kuhn RM, Smith KE, Zwieg AS. Evolutionary and biomedical insights from the rhesus macaque genome. Science. 2007;316:222–234. doi: 10.1126/science.1139247. [DOI] [PubMed] [Google Scholar]
- Goebel DJ, Poosch MS. NMDA receptor subunit gene expression in the rat brain: a quantitative analysis of endogenous mRNA levels of NR1Com, NR2A, NR2B, NR2C, NR2D and NR3A. Brain Res Mol Brain Res. 1999;69:164–170. doi: 10.1016/s0169-328x(99)00100-x. [DOI] [PubMed] [Google Scholar]
- Gonzalez MM, Valatx JL, Debilly G. Role of the locus coeruleus in the sleep rebound following two different sleep deprivation methods in the rat. Brain research. 1996;740:215–226. doi: 10.1016/s0006-8993(96)00871-2. [DOI] [PubMed] [Google Scholar]
- Gonzalez MM, Aston-Jones G. Circadian regulation of arousal: role of the noradrenergic locus coeruleus system and light exposure. Sleep. 2006;29:1327–1336. doi: 10.1093/sleep/29.10.1327. [DOI] [PubMed] [Google Scholar]
- Hollmann M, O’Shea-Greenfield A, Rogers SW, Heinemann S. Cloning by functional expression of a member of the glutamate receptor family. Nature. 1989;342:643–648. doi: 10.1038/342643a0. [DOI] [PubMed] [Google Scholar]
- Hollmann M, Heinemann S. Cloned glutamate receptors. Annu Rev Neurosci. 1994;17:31–108. doi: 10.1146/annurev.ne.17.030194.000335. [DOI] [PubMed] [Google Scholar]
- Horvath TL, Peyron C, Diano S, Ivanov A, Aston-Jones G, Kilduff TS, van Den Pol AN. Hypocretin (orexin) activation and synaptic innervation of the locus coeruleus noradrenergic system. Journal of Comparative Neurology. 1999;415:145–159. [PubMed] [Google Scholar]
- Hoshi K, Ma T, Ho IK. Precipitated kappa-opioid receptor agonist withdrawal increase glutamate in rat locus coeruleus. Eur J Pharmacol. 1996;314:301–306. doi: 10.1016/s0014-2999(96)00569-9. [DOI] [PubMed] [Google Scholar]
- Hoshi K, Ma T, Oh S, Ho IK. Increased release of excitatory amino acids in rat locus coeruleus in kappa-opioid agonist dependent rats precipitated by nor-binaltorphimine. Brain Res. 1997;753:63–68. doi: 10.1016/s0006-8993(96)01492-8. [DOI] [PubMed] [Google Scholar]
- Ishii T, Moriyoshi K, Sugihara H, Sakurada K, Kadotani H, Yokoi M, Akazawa C, Shigemoto R, Mizuno N, Masu M, et al. Molecular characterization of the family of the N-methyl-D-aspartate receptor subunits. J Biol Chem. 1993;268:2836–2843. [PubMed] [Google Scholar]
- Jodo E, Aston-Jones G. Activation of locus coeruleus by prefrontal cortex is mediated by excitatory amino acid inputs. Brain Res. 1997;768:327–332. doi: 10.1016/s0006-8993(97)00703-8. [DOI] [PubMed] [Google Scholar]
- Karolewicz B, Stockmeier CA, Ordway GA. Elevated levels of the NR2C subunit of the NMDA receptor in the locus coeruleus in depression. Neuropsychopharmacology. 2005;30:1557–1567. doi: 10.1038/sj.npp.1300781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karolewicz B, Johnson L, Szebeni K, Stockmeier CA, Ordway GA. Glutamate signaling proteins and tyrosine hydroxylase in the locus coeruleus of alcoholics. 2007 doi: 10.1016/j.jpsychires.2007.02.010. Epub ahead of print. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=17481661. [DOI] [PMC free article] [PubMed]
- Kohama SG, Urbanski HF. Distribution of glutamate receptor subunits in the primate temporal cortex and hippocampus. Brain Res. 1997;769:44–56. doi: 10.1016/s0006-8993(97)00686-0. [DOI] [PubMed] [Google Scholar]
- Kohama SG, Garyfallou VT, Urbanski HF. Regional distribution of glutamate receptor mRNA in the monkey hippocampus and temporal cortex: influence of estradiol. Brain Res Mol Brain Res. 1998;53:328–332. doi: 10.1016/s0169-328x(97)00282-9. [DOI] [PubMed] [Google Scholar]
- Kohr G. NMDA receptor function: subunit composition versus spatial distribution. Cell Tissue Res. 2006;326:439–446. doi: 10.1007/s00441-006-0273-6. [DOI] [PubMed] [Google Scholar]
- Leranth C, MacLusky NJ, Shanabrough M, Naftolin F. Catecholaminergic innervation of luteinizing hormone-releasing hormone and glutamic acid decarboxylase immunopositive neurons in the rat medial preoptic area. An electron-microscopic double immunostaining and degeneration study. Neuroendocrinology. 1988;48:591–602. doi: 10.1159/000125068. [DOI] [PubMed] [Google Scholar]
- Loughlin SE, Foote SL, Bloom FE. Efferent projections of nucleus locus coeruleus: topographic organization of cells of origin demonstrated by three-dimensional reconstruction. Neuroscience. 1986;18:291–306. doi: 10.1016/0306-4522(86)90155-7. [DOI] [PubMed] [Google Scholar]
- Luppi PH, Aston-Jones G, Akaoka H, Chouvet G, Jouvet M. Afferent projections to the rat locus coeruleus demonstrated by retrograde and anterograde tracing with cholera-toxin B subunit and Phaseolus vulgaris leucoagglutinin. Neuroscience. 1995;65:119–160. doi: 10.1016/0306-4522(94)00481-j. [DOI] [PubMed] [Google Scholar]
- Luque JM, Malherbe P, Richards JG. Localization of NMDA receptor subunit mRNAs in the rat locus coeruleus. Brain Res Mol Brain Res. 1995;29:224–232. doi: 10.1016/0169-328x(94)00253-b. [DOI] [PubMed] [Google Scholar]
- Mason ST, Fibiger HC. Regional topography within noradrenergic locus coeruleus as revealed by retrograde transport of horseradish peroxidase. Journal of comparative neurology. 1979;187:703–724. doi: 10.1002/cne.901870405. [DOI] [PubMed] [Google Scholar]
- Medhamurthy R, Gay VL, Plant TM. Repetitive injections of L-glutamic acid, in contrast to those of N-methyl-D,L-aspartic acid, fail to elicit sustained hypothalamic GnRH release in the prepubertal male rhesus monkey (Macaca mulatta) Neuroendocrinology. 1992;55:660–666. doi: 10.1159/000126186. [DOI] [PubMed] [Google Scholar]
- Meguro H, Mori H, Araki K, Kushiya E, Kutsuwada T, Yamazaki M, Kumanishi T, Arakawa M, Sakimura K, Mishina M. Functional characterization of a heteromeric NMDA receptor channel expressed from cloned cDNAs. Nature. 1992;357:70–74. doi: 10.1038/357070a0. [DOI] [PubMed] [Google Scholar]
- Monyer H, Sprengel R, Schoepfer R, Herb A, Higuchi M, Lomeli H, Burnashev N, Sakmann B, Seeburg PH. Heteromeric NMDA receptors: molecular and functional distinction of subtypes. Science. 1992;256:1217–1221. doi: 10.1126/science.256.5060.1217. [DOI] [PubMed] [Google Scholar]
- Monyer H, Burnashev N, Laurie DJ, Sakmann B, Seeburg PH. Developmental and regional expression in the rat brain and functional properties of four NMDA receptors. Neuron. 1994;12:529–540. doi: 10.1016/0896-6273(94)90210-0. [DOI] [PubMed] [Google Scholar]
- Moore RY, Bloom FE. Central catecholamine neuron systems: anatomy and physiology of the norepinephrine and epinephrine systems. Annu Rev Neurosci. 1979;2:113–168. doi: 10.1146/annurev.ne.02.030179.000553. [DOI] [PubMed] [Google Scholar]
- Moriyoshi K, Masu M, Ishii T, Shigemoto R, Mizuno N, Nakanishi S. Molecular cloning and characterization of the rat NMDA receptor. Nature. 1991;354:31–37. doi: 10.1038/354031a0. [DOI] [PubMed] [Google Scholar]
- Nakanishi S. Molecular diversity of glutamate receptors and implications for brain function. Science. 1992;258:597–603. doi: 10.1126/science.1329206. [DOI] [PubMed] [Google Scholar]
- Nakanishi S, Masu M. Molecular diversity and functions of glutamate receptors. Annu Rev Biophys Biomol Struct. 1994;23:319–348. doi: 10.1146/annurev.bb.23.060194.001535. [DOI] [PubMed] [Google Scholar]
- Nakanishi S, Masu M, Bessho Y, Nakajima Y, Hayashi Y, Shigemoto R. Molecular diversity of glutamate receptors and their physiological functions. Exs. 1994;71:71–80. doi: 10.1007/978-3-0348-7330-7_8. [DOI] [PubMed] [Google Scholar]
- Nitz D, Siegel JM. GABA release in the locus coeruleus as a function of sleep/wake state. Neuroscience. 1997;78:795–801. doi: 10.1016/s0306-4522(96)00549-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Neill MJ, Bleakman D, Zimmerman DM, Nisenbaum ES. AMPA receptor potentiators for the treatment of CNS disorders. Curr Drug Targets CNS Neurol Disord. 2004;3:181–194. doi: 10.2174/1568007043337508. [DOI] [PubMed] [Google Scholar]
- Olson CR, Freeman RD. Profile of the sensitive period for monocular deprivation in kittens. Exp Brain Res. 1980;39:17–21. doi: 10.1007/BF00237065. [DOI] [PubMed] [Google Scholar]
- Palucha A, Pilc A. The involvement of glutamate in the pathophysiology of depression. Drug News Perspect. 2005;18:262–268. doi: 10.1358/dnp.2005.18.4.908661. [DOI] [PubMed] [Google Scholar]
- Pau KY, Hess DL, Kaynard AH, Ji WZ, Gliessman PM, Spies HG. Suppression of mediobasal hypothalamic gonadotropin-releasing hormone and plasma luteinizing hormone pulsatile patterns by phentolamine in ovariectomized rhesus macaques. Endocrinology. 1989;124:891–898. doi: 10.1210/endo-124-2-891. [DOI] [PubMed] [Google Scholar]
- Pau KY, Ma YJ, Yu JH, Yang SP, Airhart N, Spies HG. Topographic comparison of the expression of norepinephrine transporter, tyrosine hydroxylase and neuropeptide Y mRNA in association with dopamine beta-hydroxylase neurons in the rabbit brainstem. Brain research. 1997;48:367–381. doi: 10.1016/s0169-328x(97)00113-7. [DOI] [PubMed] [Google Scholar]
- Pau KY, Lee CJ, Cowles A, Yang SP, Hess DL, Spies HG. Possible involvement of norepinephrine transporter activity in the pulsatility of hypothalamic gonadotropin-releasing hormone release: influence of the gonad. Journal of neuroendocrinology. 1998;10:21–29. doi: 10.1046/j.1365-2826.1998.00173.x. [DOI] [PubMed] [Google Scholar]
- Pau KY, Hess DL, Kohama S, Bao J, Pau CY, Spies HG. Oestrogen upregulates noradrenaline release in the mediobasal hypothalamus and tyrosine hydroxylase gene expression in the brainstem of ovariectomized rhesus macaques. J Neuroendocrinol. 2000;12:899–909. doi: 10.1046/j.1365-2826.2000.00549.x. [DOI] [PubMed] [Google Scholar]
- Petralia RS, Wenthold RJ. Light and electron immunocytochemical localization of AMPA-selective glutamate receptors in the rat brain. J Comp Neurol. 1992;318:329–354. doi: 10.1002/cne.903180309. [DOI] [PubMed] [Google Scholar]
- Petralia RS, Yokotani N, Wenthold RJ. Light and electron microscope distribution of the NMDA receptor subunit NMDAR1 in the rat nervous system using a selective anti-peptide antibody. J Neurosci. 1994;14:667–696. doi: 10.1523/JNEUROSCI.14-02-00667.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Planells-Cases R, Lerma J, Ferrer-Montiel A. Pharmacological intervention at ionotropic glutamate receptor complexes. Curr Pharm Des. 2006;12:3583–3596. doi: 10.2174/138161206778522092. [DOI] [PubMed] [Google Scholar]
- Plant TM, Gay VL, Marshall GR, Arslan M. Puberty in monkeys is triggered by chemical stimulation of the hypothalamus. Proceedings of the National Academy of Sciences of the United States of America. 1989;86:2506–2510. doi: 10.1073/pnas.86.7.2506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porkka-Heiskanen T, Smith SE, Taira T, Urban JH, Levine JE, Turek FW, Stenberg D. Noradrenergic activity in rat brain during rapid eye movement sleep deprivation and rebound sleep. Am J Physiol. 1995;268:R1456–1463. doi: 10.1152/ajpregu.1995.268.6.R1456. [DOI] [PubMed] [Google Scholar]
- Rajkowski J, Majczynski H, Clayton E, Aston-Jones G. Activation of monkey locus coeruleus neurons varies with difficulty and performance in a target detection task. J Neurophysiol. 2004;92:361–371. doi: 10.1152/jn.00673.2003. [DOI] [PubMed] [Google Scholar]
- Rasmussen K. The role of the locus coeruleus and N-methyl-D-aspartic acid (NMDA) and AMPA receptors in opiate withdrawal. Neuropsychopharmacology. 1995;13:295–300. doi: 10.1016/0893-133X(95)00082-O. [DOI] [PubMed] [Google Scholar]
- Rasmussen K, Kendrick WT, Kogan JH, Aghajanian GK. A selective AMPA antagonist, LY293558, suppresses morphine withdrawal-induced activation of locus coeruleus neurons and behavioral signs of morphine withdrawal. Neuropsychopharmacology. 1996;15:497–505. doi: 10.1016/S0893-133X(96)00094-2. [DOI] [PubMed] [Google Scholar]
- Rasmussen K, Hsu MA, Vandergriff J. The selective mGlu2/3 receptor antagonist LY341495 exacerbates behavioral signs of morphine withdrawal and morphine-withdrawal-induced activation of locus coeruleus neurons. Neuropharmacology. 2004;46:620–628. doi: 10.1016/j.neuropharm.2003.11.013. [DOI] [PubMed] [Google Scholar]
- Rasmussen K, Martin H, Berger JE, Seager MA. The mGlu5 receptor antagonists MPEP and MTEP attenuate behavioral signs of morphine withdrawal and morphine-withdrawal-induced activation of locus coeruleus neurons in rats. Neuropharmacology. 2005;48:173–180. doi: 10.1016/j.neuropharm.2004.09.010. [DOI] [PubMed] [Google Scholar]
- Reyes A, Xia LN, Ferin M. Modulation of the effects of N-methyl-D,L-aspartate on luteinizing hormone by the ovarian steroids in the adult rhesus monkey. Neuroendocrinology. 1991;54:405–411. doi: 10.1159/000125921. [DOI] [PubMed] [Google Scholar]
- Rigby M, Le Bourdelles B, Heavens RP, Kelly S, Smith D, Butler A, Hammans R, Hills R, Xuereb JH, Hill RG, Whiting PJ, Sirinathsinghji DJ. The messenger RNAs for the N-methyl-D-aspartate receptor subunits show region-specific expression of different subunit composition in the human brain. Neuroscience. 1996;73:429–447. doi: 10.1016/0306-4522(96)00089-9. [DOI] [PubMed] [Google Scholar]
- Sawchenko PE, Swanson LW. Central noradrenergic pathways for the integration of hypothalamic neuroendocrine and autonomic responses. Science. 1981;214:685–687. doi: 10.1126/science.7292008. [DOI] [PubMed] [Google Scholar]
- Sawchenko PE, Swanson LW. The organization of noradrenergic pathways from the brainstem to the paraventricular and supraoptic nuclei in the rat. Brain research. 1982;257:275–325. doi: 10.1016/0165-0173(82)90010-8. [DOI] [PubMed] [Google Scholar]
- Sawchenko PE, Swanson LW, Grzanna R, Howe PR, Bloom SR, Polak JM. Colocalization of neuropeptide Y immunoreactivity in brainstem catecholaminergic neurons that project to the paraventricular nucleus of the hypothalamus. Journal of comparative neurology. 1985;241:138–153. doi: 10.1002/cne.902410203. [DOI] [PubMed] [Google Scholar]
- Singewald N, Schneider C, Philippu A. Effects of neuroactive compounds, noxious and cardiovascular stimuli on the release of amino acids in the rat locus coeruleus. Neurosci Lett. 1994;180:55–58. doi: 10.1016/0304-3940(94)90912-1. [DOI] [PubMed] [Google Scholar]
- Singewald N, Zhou GY, Schneider C. Release of excitatory and inhibitory amino acids from the locus coeruleus of conscious rats by cardiovascular stimuli and various forms of acute stress. Brain Res. 1995;704:42–50. doi: 10.1016/0006-8993(95)01102-1. [DOI] [PubMed] [Google Scholar]
- Singewald N, Zhou GY, Chen F, Philippu A. Corticotropin-releasing factor modulates basal and stress-induced excitatory amino acid release in the locus coeruleus of conscious rats. Neurosci Lett. 1996;204:45–48. doi: 10.1016/0304-3940(96)12312-0. [DOI] [PubMed] [Google Scholar]
- Singewald N, Philippu A. Release of neurotransmitters in the locus coeruleus. Prog Neurobiol. 1998;56:237–267. doi: 10.1016/s0301-0082(98)00039-2. [DOI] [PubMed] [Google Scholar]
- Sommer B, Seeburg PH. Glutamate receptor channels: novel properties and new clones. Trends Pharmacol Sci. 1992;13:291–296. doi: 10.1016/0165-6147(92)90088-n. [DOI] [PubMed] [Google Scholar]
- Spies HG, Pau KY, Yang SP. Coital and estrogen signals: a contrast in the preovulatory neuroendocrine networks of rabbits and rhesus monkeys. Biology of reproduction. 1997;56:310–319. doi: 10.1095/biolreprod56.2.310. [DOI] [PubMed] [Google Scholar]
- Sucher NJ, Akbarian S, Chi CL, Leclerc CL, Awobuluyi M, Deitcher DL, Wu MK, Yuan JP, Jones EG, Lipton SA. Developmental and regional expression pattern of a novel NMDA receptor-like subunit (NMDAR-L) in the rodent brain. J Neurosci. 1995;15:6509–6520. doi: 10.1523/JNEUROSCI.15-10-06509.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki T, Higgins PJ, Crawford DR. Control selection for RNA quantitation. BioTechniques. 2000;29:332–337. doi: 10.2144/00292rv02. [DOI] [PubMed] [Google Scholar]
- Takamori S, Rhee JS, Rosenmund C, Jahn R. Identification of a vesicular glutamate transporter that defines a glutamatergic phenotype in neurons. Nature. 2000;407:189–194. doi: 10.1038/35025070. [DOI] [PubMed] [Google Scholar]
- Takamori S. VGLUTs: ‘exciting’ times for glutamatergic research? Neurosci Res. 2006;55:343–351. doi: 10.1016/j.neures.2006.04.016. [DOI] [PubMed] [Google Scholar]
- Terasawa E, Krook C, Hei DL, Gearing M, Schultz NJ, Davis GA. Norepinephrine is a possible neurotransmitter stimulating pulsatile release of luteinizing hormone-releasing hormone in the rhesus monkey. Endocrinology. 1988;123:1808–1816. doi: 10.1210/endo-123-4-1808. [DOI] [PubMed] [Google Scholar]
- Tohyama M, Oyamada H. Gene expression of neuroreceptors in the locus coeruleus of the rat. Microsc Res Tech. 1994;29:200–203. doi: 10.1002/jemt.1070290304. [DOI] [PubMed] [Google Scholar]
- Tokuyama S, Ho IK. Inhibitory effects of diltiazem, an L-type Ca2+ channel blocker, on naloxone-increased glutamate levels in the locus coeruleus of opioid-dependent rats. Brain Res. 1996;722:212–216. doi: 10.1016/0006-8993(96)00187-4. [DOI] [PubMed] [Google Scholar]
- Urbanski HF, Kohama SG, Garyfallou VT. Mechanisms mediating the response of GnRH neurones to excitatory amino acids. Rev Reprod. 1996;1:173–181. doi: 10.1530/ror.0.0010173. [DOI] [PubMed] [Google Scholar]
- Urbanski HF, Garyfallou VT, Kohama SG, Hess DL. Alpha-adrenergic receptor antagonism and N-methyl-D-aspartate (NMDA) induced luteinizing hormone release in female rhesus macaques. Brain Res. 1997;744:96–104. doi: 10.1016/s0006-8993(96)01083-9. [DOI] [PubMed] [Google Scholar]
- Vallano ML. Developmental aspects of NMDA receptor function. Crit Rev Neurobiol. 1998;12:177–204. doi: 10.1615/critrevneurobiol.v12.i3.20. [DOI] [PubMed] [Google Scholar]
- Vandergriff J, Rasmussen K. The selective mGlu2/3 receptor agonist LY354740 attenuates morphine-withdrawal-induced activation of locus coeruleus neurons and behavioral signs of morphine withdrawal. Neuropharmacology. 1999;38:217–222. doi: 10.1016/s0028-3908(98)00196-8. [DOI] [PubMed] [Google Scholar]
- Wafford KA, Bain CJ, Le Bourdelles B, Whiting PJ, Kemp JA. Preferential co-assembly of recombinant NMDA receptors composed of three different subunits. Neuroreport. 1993;4:1347–1349. doi: 10.1097/00001756-199309150-00015. [DOI] [PubMed] [Google Scholar]
- Walling SG, Harley CW. Locus ceruleus activation initiates delayed synaptic potentiation of perforant path input to the dentate gyrus in awake rats: a novel beta-adrenergic- and protein synthesis-dependent mammalian plasticity mechanism. J Neurosci. 2004;24:598–604. doi: 10.1523/JNEUROSCI.4426-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenthold RJ, Yokotani N, Doi K, Wada K. Immunochemical characterization of the non-NMDA glutamate receptor using subunit-specific antibodies. Evidence for a hetero-oligomeric structure in rat brain. J Biol Chem. 1992;267:501–507. [PubMed] [Google Scholar]
- Wilson RC, Knobil E. Acute effects of N-methyl-DL-aspartate on the release of pituitary gonadotropins and prolactin in the adult female rhesus monkey. Brain research. 1982;248:177–179. doi: 10.1016/0006-8993(82)91160-x. [DOI] [PubMed] [Google Scholar]
- Wu MF, Gulyani SA, Yau E, Mignot E, Phan B, Siegel JM. Locus coeruleus neurons: cessation of activity during cataplexy. Neuroscience. 1999;91:1389–1399. doi: 10.1016/s0306-4522(98)00600-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang SP, Pau KY, Spies HG. Tyrosine hydroxylase and norepinephrine transporter mRNA levels increase in locus coeruleus after coitus in rabbits. Journal of molecular endocrinology. 1997;19:311–319. doi: 10.1677/jme.0.0190311. [DOI] [PubMed] [Google Scholar]
- Yang SP, Pau KY, Spies HG. Gonadectomy alters tyrosine hydroxylase and norepinephrine transporter mRNA levels in the locus coeruleus in rabbits. Journal of neuroendocrinology. 1997;9:763–768. doi: 10.1046/j.1365-2826.1997.00641.x. [DOI] [PubMed] [Google Scholar]
- Yang SP, Pau KY, Airhart N, Spies HG. Attenuation of gonadotropin-releasing hormone reflex to coitus by alpha1-adrenergic receptor blockade in the rabbit. Proceedings of the Society for Experimental Biology and Medicine. 1998;218:204–209. doi: 10.3181/00379727-218-44287. [DOI] [PubMed] [Google Scholar]
- Zhang T, Feng Y, Rockhold RW, Ho IK. Naloxone-precipitated morphine withdrawal increases pontine glutamate levels in the rat. Life Sci. 1994;55:PL25–31. doi: 10.1016/0024-3205(94)90108-2. [DOI] [PubMed] [Google Scholar]
- Zukin RS, Bennett MV. Alternatively spliced isoforms of the NMDARI receptor subunit. Trends Neurosci. 1995;18:306–313. doi: 10.1016/0166-2236(95)93920-s. [DOI] [PubMed] [Google Scholar]


