Abstract
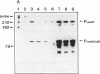
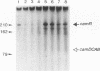
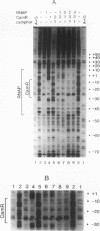
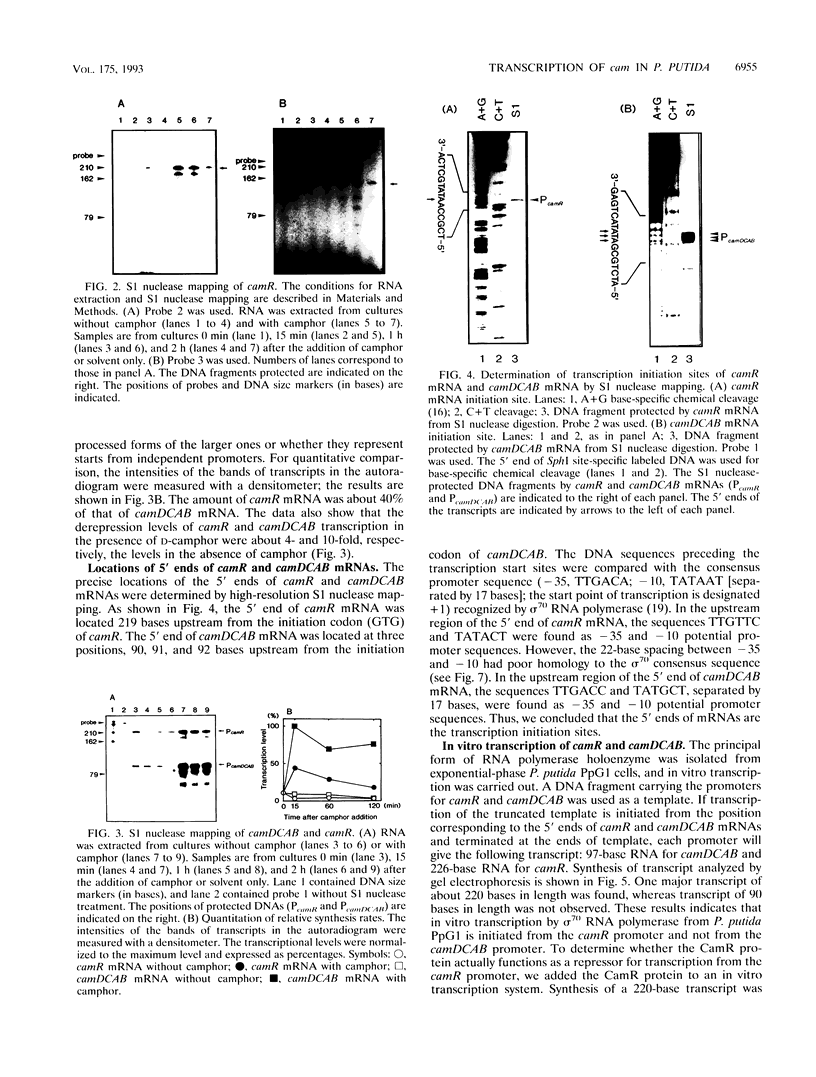
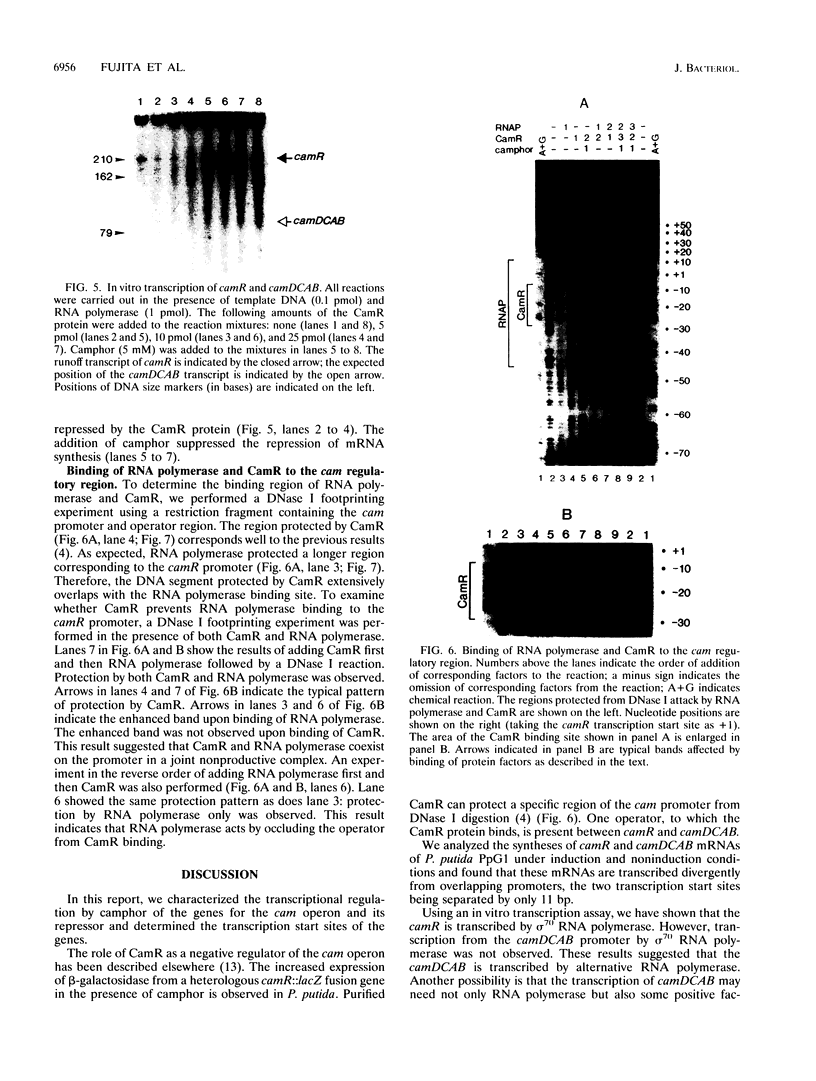
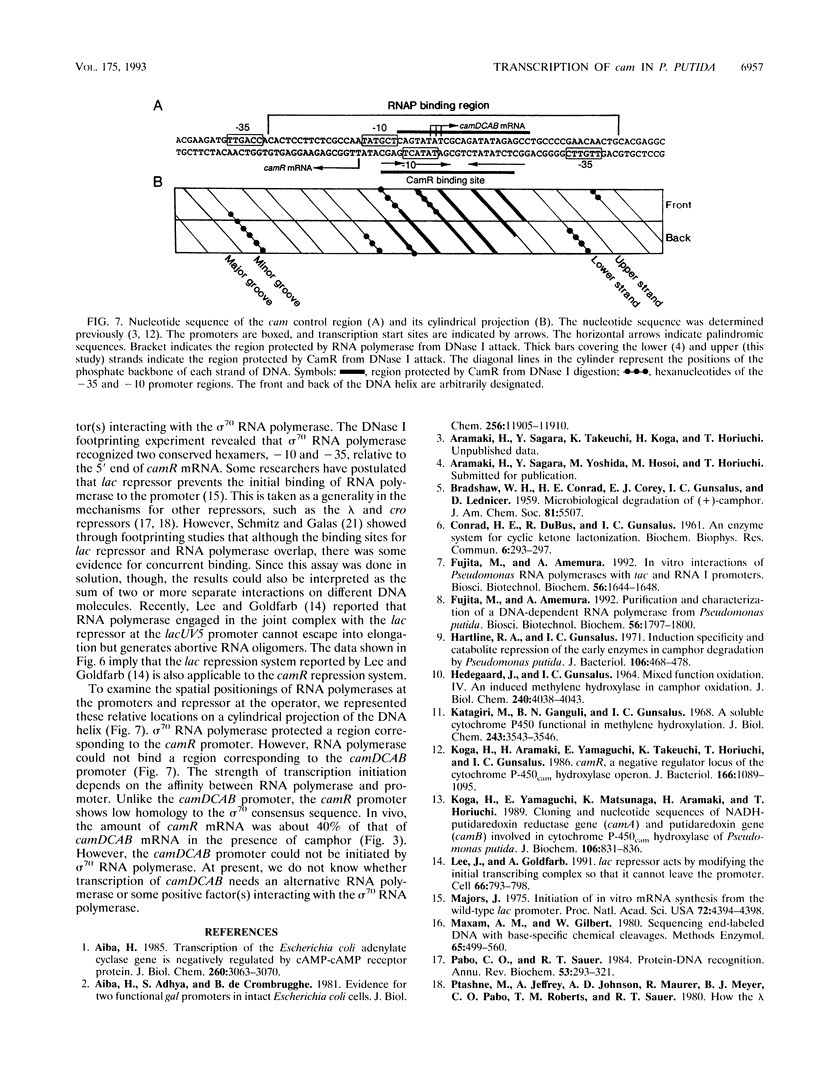
In Pseudomonas putida carrying the CAM plasmid, the operon (camDCAB) encoding enzymes involved in the degradation pathway of D-camphor is negatively regulated by the CamR protein, and camR is autorepressed. S1 nuclease mapping revealed that camDCAB and camR were divergently transcribed from overlapping promoters, the transcription start sites were separated by 11 bp, and transcriptions of the cam operon (camDCAB) and camR increased about 10- and 4-fold, respectively, immediately after addition of camphor. The transcriptions of camDCAB and camR were negatively regulated through the interaction of the CamR protein with the one operator located in the overlapping promoter region. In vitro transcription experiments were performed to characterize the regulation of cam genes. The camR promoter was initiated by P. putida RNA polymerase containing sigma 70, but transcription from the camDCAB promoter by sigma 70 holoenzyme was not observed. The purified CamR protein repressed in vitro transcription from the camR promoter. This repression was suppressed by camphor. The RNA polymerase binding region of the camR promoter was identified by using DNase I footprinting. In addition, footprinting studies revealed that the CamR protein and RNA polymerase coexisted on the promoter region in a joint nonproductive complex.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Aiba H., Adhya S., de Crombrugghe B. Evidence for two functional gal promoters in intact Escherichia coli cells. J Biol Chem. 1981 Nov 25;256(22):11905–11910. [PubMed] [Google Scholar]
- Aiba H. Transcription of the Escherichia coli adenylate cyclase gene is negatively regulated by cAMP-cAMP receptor protein. J Biol Chem. 1985 Mar 10;260(5):3063–3070. [PubMed] [Google Scholar]
- CONRAD H. E., DUBUS R., GUNSALUS I. C. An enzyme system for cyclic ketone lactonization. Biochem Biophys Res Commun. 1961 Nov 29;6:293–297. doi: 10.1016/0006-291x(61)90382-5. [DOI] [PubMed] [Google Scholar]
- Fujita M., Amemura A. In vitro interactions of Pseudomonas RNA polymerases with tac and RNA I promoters. Biosci Biotechnol Biochem. 1992 Oct;56(10):1644–1648. doi: 10.1271/bbb.56.1644. [DOI] [PubMed] [Google Scholar]
- Fujita M., Amemura A. Purification and characterization of a DNA-dependent RNA polymerase from Pseudomonas putida. Biosci Biotechnol Biochem. 1992 Nov;56(11):1797–1800. doi: 10.1271/bbb.56.1797. [DOI] [PubMed] [Google Scholar]
- Hartline R. A., Gunsalus I. C. Induction specificity and catabolite repression of the early enzymes in camphor degradation by Pseudomonas putida. J Bacteriol. 1971 May;106(2):468–478. doi: 10.1128/jb.106.2.468-478.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedegaard J., Gunsalus I. C. Mixed function oxidation. IV. An induced methylene hydroxylase in camphor oxidation. J Biol Chem. 1965 Oct;240(10):4038–4043. [PubMed] [Google Scholar]
- Katagiri M., Ganguli B. N., Gunsalus I. C. A soluble cytochrome P-450 functional in methylene hydroxylation. J Biol Chem. 1968 Jun 25;243(12):3543–3546. [PubMed] [Google Scholar]
- Koga H., Aramaki H., Yamaguchi E., Takeuchi K., Horiuchi T., Gunsalus I. C. camR, a negative regulator locus of the cytochrome P-450cam hydroxylase operon. J Bacteriol. 1986 Jun;166(3):1089–1095. doi: 10.1128/jb.166.3.1089-1095.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koga H., Yamaguchi E., Matsunaga K., Aramaki H., Horiuchi T. Cloning and nucleotide sequences of NADH-putidaredoxin reductase gene (camA) and putidaredoxin gene (camB) involved in cytochrome P-450cam hydroxylase of Pseudomonas putida. J Biochem. 1989 Nov;106(5):831–836. doi: 10.1093/oxfordjournals.jbchem.a122939. [DOI] [PubMed] [Google Scholar]
- Lee J., Goldfarb A. lac repressor acts by modifying the initial transcribing complex so that it cannot leave the promoter. Cell. 1991 Aug 23;66(4):793–798. doi: 10.1016/0092-8674(91)90122-f. [DOI] [PubMed] [Google Scholar]
- Majors J. Initiation of in vitro mRNA synthesis from the wild-type lac promoter. Proc Natl Acad Sci U S A. 1975 Nov;72(11):4394–4398. doi: 10.1073/pnas.72.11.4394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Pabo C. O., Sauer R. T. Protein-DNA recognition. Annu Rev Biochem. 1984;53:293–321. doi: 10.1146/annurev.bi.53.070184.001453. [DOI] [PubMed] [Google Scholar]
- Reznikoff W. S., Siegele D. A., Cowing D. W., Gross C. A. The regulation of transcription initiation in bacteria. Annu Rev Genet. 1985;19:355–387. doi: 10.1146/annurev.ge.19.120185.002035. [DOI] [PubMed] [Google Scholar]
- Rheinwald J. G., Chakrabarty A. M., Gunsalus I. C. A transmissible plasmid controlling camphor oxidation in Pseudomonas putida. Proc Natl Acad Sci U S A. 1973 Mar;70(3):885–889. doi: 10.1073/pnas.70.3.885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz A., Galas D. J. The interaction of RNA polymerase and lac repressor with the lac control region. Nucleic Acids Res. 1979 Jan;6(1):111–137. doi: 10.1093/nar/6.1.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ullah A. J., Murray R. I., Bhattacharyya P. K., Wagner G. C., Gunsalus I. C. Protein components of a cytochrome P-450 linalool 8-methyl hydroxylase. J Biol Chem. 1990 Jan 25;265(3):1345–1351. [PubMed] [Google Scholar]