Abstract
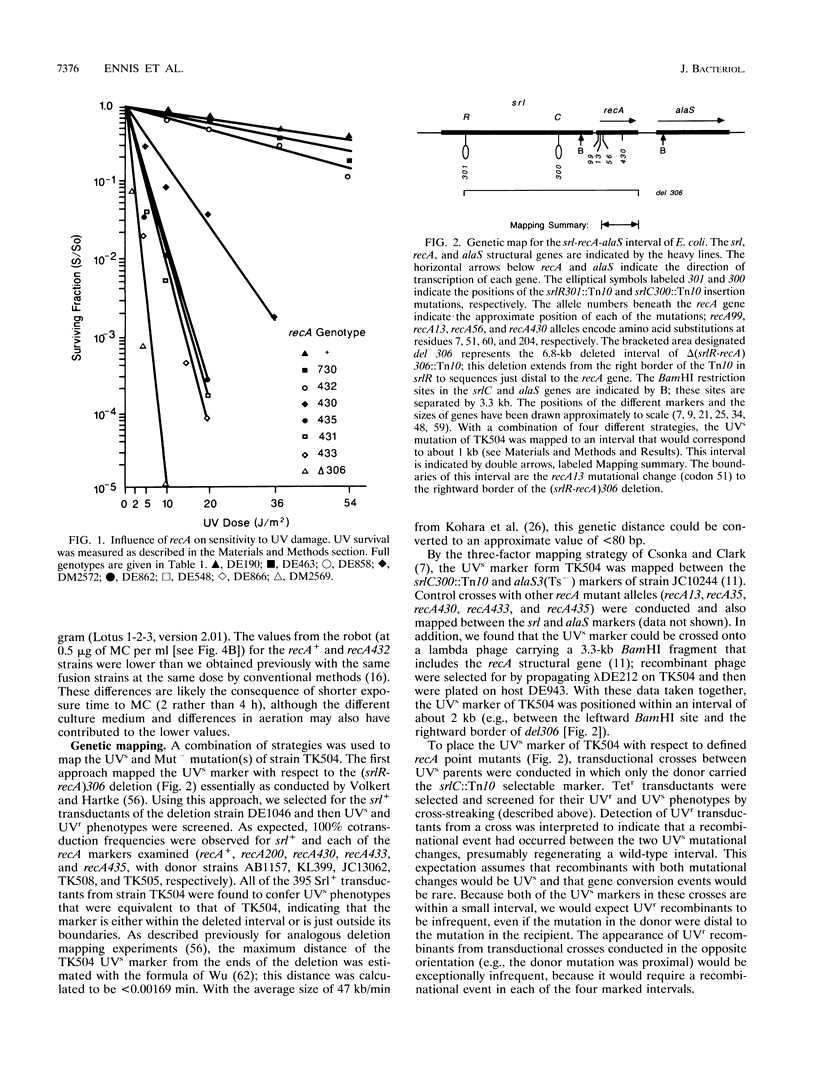
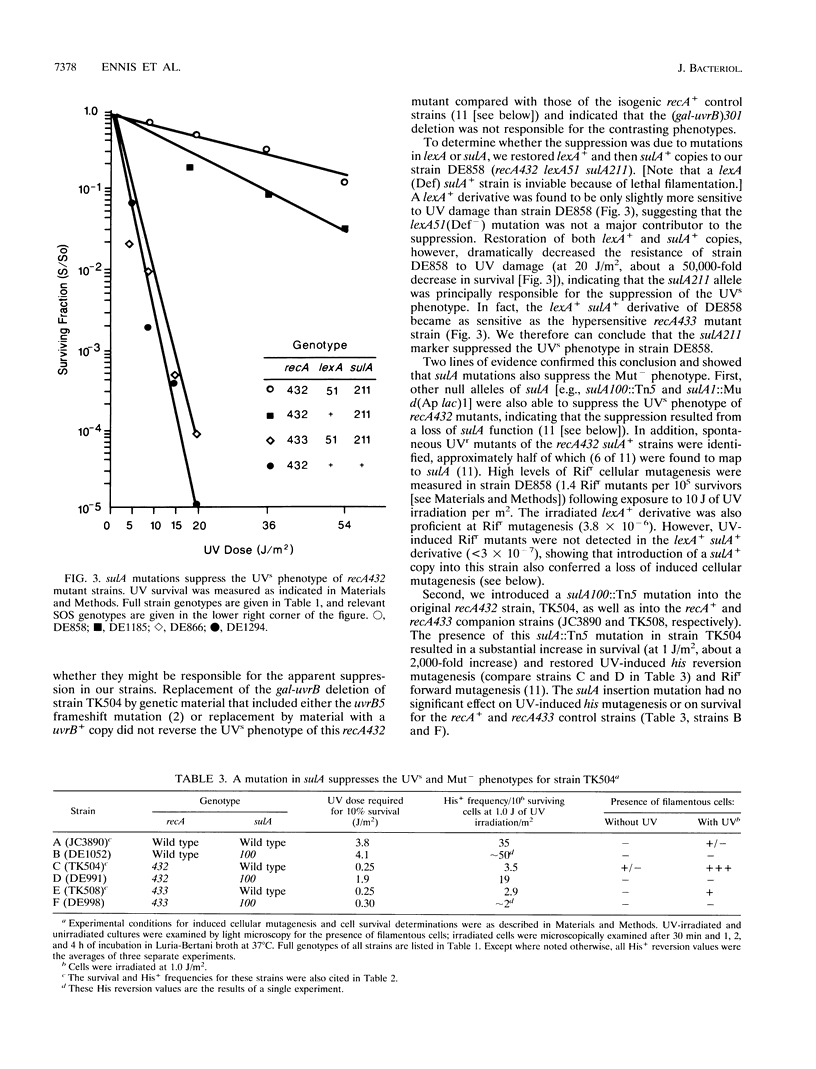
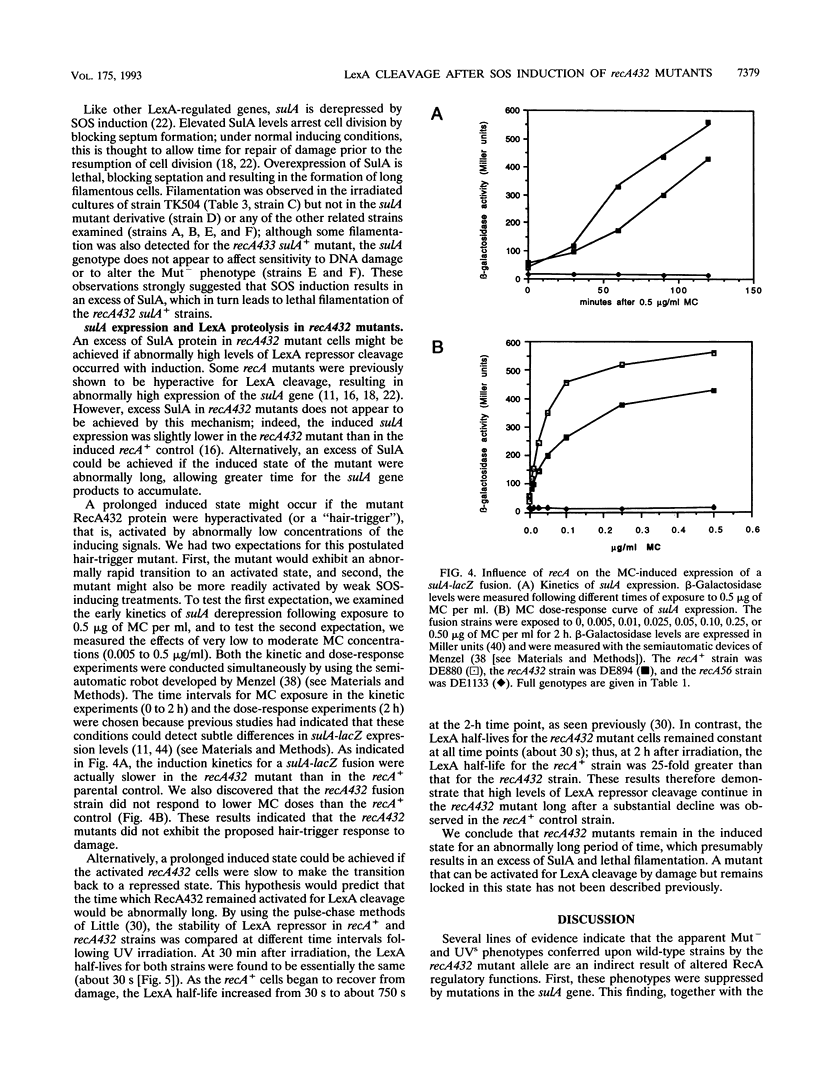
The recA432 mutant allele was isolated (T. Kato and Y. Shinoura, Mol. Gen. Genet. 156:121-131, 1977) by virtue of its defect in cellular mutagenesis (Mut-) and its hypersensitivity to damage by UV irradiation (UVs), which were phenotypes expected for a recA mutant. However, we found that in a different genetic background (lexA51 sulA211 uvrB+), recA432 mutants expressed certain mutant phenotypes but not the Mut- and UVs phenotypes (D.G. Ennis, N. Ossanna, and D.W. Mount, J. Bacteriol. 171:2533-2541, 1989). We present several lines of evidence that these differences resulted from the sulA genotype of the cell and that the apparent UVs and Mut- phenotypes of the sulA+ derivatives resulted from lethal filamentation of induced cells because of persistent derepression of sulA. First, transduction of sulA(Def) mutations into the recA432 strains restored cellular mutagenesis and resistance to UV. Second, recA432 sulA+ strains underwent filamentous death following SOS-inducing treatments. Third, cleavage of LexA repressor in a recA432 strain continued at a rapid rate long after UV induction, at a time when cleavage of the repressor in the recA+ parental strain had substantially declined. Fourth, we confirmed that a single mutation (recA432) conferring both the UVs and Mut- phenotypes mapped to the recA gene. These findings indicate that the RecA432 mutant protein is defective in making the transition back to the deactivated state following SOS induction; thus, the SOS-induced state of recA432 mutants is prolonged and can account for an excess of SulA protein, leading to filamentation. These results are discussed in the context of molecular models for RecA activation for LexA and UmuD cleavage and their roles in the control of mutagenesis and cell division in the SOS response.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Backendorf C., Spaink H., Barbeiro A. P., van de Putte P. Structure of the uvrB gene of Escherichia coli. Homology with other DNA repair enzymes and characterization of the uvrB5 mutation. Nucleic Acids Res. 1986 Apr 11;14(7):2877–2890. doi: 10.1093/nar/14.7.2877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanco M., Herrera G., Collado P., Rebollo J. E., Botella L. M. Influence of RecA protein on induced mutagenesis. Biochimie. 1982 Aug-Sep;64(8-9):633–636. doi: 10.1016/s0300-9084(82)80102-8. [DOI] [PubMed] [Google Scholar]
- Burckhardt S. E., Woodgate R., Scheuermann R. H., Echols H. UmuD mutagenesis protein of Escherichia coli: overproduction, purification, and cleavage by RecA. Proc Natl Acad Sci U S A. 1988 Mar;85(6):1811–1815. doi: 10.1073/pnas.85.6.1811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caillet-Fauquet P., Maenhaut-Michel G. Nature of the SOS mutator activity: genetic characterization of untargeted mutagenesis in Escherichia coli. Mol Gen Genet. 1988 Aug;213(2-3):491–498. doi: 10.1007/BF00339621. [DOI] [PubMed] [Google Scholar]
- Clark A. J. recA operator mutations and their usefulness. Biochimie. 1982 Aug-Sep;64(8-9):669–675. doi: 10.1016/s0300-9084(82)80108-9. [DOI] [PubMed] [Google Scholar]
- Csonka L. N., Clark A. J. Deletions generated by the transposon Tn10 in the srl recA region of the Escherichia coli K-12 chromosome. Genetics. 1979 Oct;93(2):321–343. doi: 10.1093/genetics/93.2.321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutreix M., Burnett B., Bailone A., Radding C. M., Devoret R. A partially deficient mutant, recA1730, that fails to form normal nucleoprotein filaments. Mol Gen Genet. 1992 Apr;232(3):489–497. doi: 10.1007/BF00266254. [DOI] [PubMed] [Google Scholar]
- Dutreix M., Moreau P. L., Bailone A., Galibert F., Battista J. R., Walker G. C., Devoret R. New recA mutations that dissociate the various RecA protein activities in Escherichia coli provide evidence for an additional role for RecA protein in UV mutagenesis. J Bacteriol. 1989 May;171(5):2415–2423. doi: 10.1128/jb.171.5.2415-2423.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisen H., Brachet P., Pereira da Silva L., Jacob F. Regulation of repressor expression in lambda. Proc Natl Acad Sci U S A. 1970 Jul;66(3):855–862. doi: 10.1073/pnas.66.3.855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ennis D. G., Amundsen S. K., Smith G. R. Genetic functions promoting homologous recombination in Escherichia coli: a study of inversions in phage lambda. Genetics. 1987 Jan;115(1):11–24. doi: 10.1093/genetics/115.1.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ennis D. G., Fisher B., Edmiston S., Mount D. W. Dual role for Escherichia coli RecA protein in SOS mutagenesis. Proc Natl Acad Sci U S A. 1985 May;82(10):3325–3329. doi: 10.1073/pnas.82.10.3325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ennis D. G., Ossanna N., Mount D. W. Genetic separation of Escherichia coli recA functions for SOS mutagenesis and repressor cleavage. J Bacteriol. 1989 May;171(5):2533–2541. doi: 10.1128/jb.171.5.2533-2541.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ennis D. G., Peterson K. R., Mount D. W. Increased expression of the Escherichia coli umuDC operon restores SOS mutagenesis in lexA41 cells. Mol Gen Genet. 1988 Aug;213(2-3):541–544. doi: 10.1007/BF00339628. [DOI] [PubMed] [Google Scholar]
- Glickman W., Guijt N., Morand P. The genetic characterization of lexB32, lexB33 and lexB35 mutations of Escherichia coli: location and complementation pattern for UV resistance. Mol Gen Genet. 1977 Nov 29;157(1):83–89. doi: 10.1007/BF00268690. [DOI] [PubMed] [Google Scholar]
- Hauser J., Levine A. S., Ennis D. G., Chumakov K. M., Woodgate R. The enhanced mutagenic potential of the MucAB proteins correlates with the highly efficient processing of the MucA protein. J Bacteriol. 1992 Nov;174(21):6844–6851. doi: 10.1128/jb.174.21.6844-6851.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horii T., Ogawa T., Ogawa H. Organization of the recA gene of Escherichia coli. Proc Natl Acad Sci U S A. 1980 Jan;77(1):313–317. doi: 10.1073/pnas.77.1.313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huisman O., D'Ari R. An inducible DNA replication-cell division coupling mechanism in E. coli. Nature. 1981 Apr 30;290(5809):797–799. doi: 10.1038/290797a0. [DOI] [PubMed] [Google Scholar]
- Kato T., Rothman R. H., Clark A. J. Analysis of the role of recombination and repair in mutagenesis of Escherichia coli by UV irradiation. Genetics. 1977 Sep;87(1):1–18. doi: 10.1093/genetics/87.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato T., Shinoura Y. Isolation and characterization of mutants of Escherichia coli deficient in induction of mutations by ultraviolet light. Mol Gen Genet. 1977 Nov 14;156(2):121–131. doi: 10.1007/BF00283484. [DOI] [PubMed] [Google Scholar]
- Kawashima H., Horii T., Ogawa T., Ogawa H. Functional domains of Escherichia coli recA protein deduced from the mutational sites in the gene. Mol Gen Genet. 1984;193(2):288–292. doi: 10.1007/BF00330682. [DOI] [PubMed] [Google Scholar]
- Kohara Y., Akiyama K., Isono K. The physical map of the whole E. coli chromosome: application of a new strategy for rapid analysis and sorting of a large genomic library. Cell. 1987 Jul 31;50(3):495–508. doi: 10.1016/0092-8674(87)90503-4. [DOI] [PubMed] [Google Scholar]
- Kowalczykowski S. C. Biochemical and biological function of Escherichia coli RecA protein: behavior of mutant RecA proteins. Biochimie. 1991 Feb-Mar;73(2-3):289–304. doi: 10.1016/0300-9084(91)90216-n. [DOI] [PubMed] [Google Scholar]
- Lavery P. E., Kowalczykowski S. C. Biochemical basis of the constitutive repressor cleavage activity of recA730 protein. A comparison to recA441 and recA803 proteins. J Biol Chem. 1992 Oct 15;267(29):20648–20658. [PubMed] [Google Scholar]
- Lieberman H. B., Witkin E. M. DNA degradation, UV sensitivity and SOS-mediated mutagenesis in strains of Escherichia coli deficient in single-strand DNA binding protein: effects of mutations and treatments that alter levels of Exonuclease V or recA protein. Mol Gen Genet. 1983;190(1):92–100. doi: 10.1007/BF00330329. [DOI] [PubMed] [Google Scholar]
- Little J. W., Mount D. W. The SOS regulatory system of Escherichia coli. Cell. 1982 May;29(1):11–22. doi: 10.1016/0092-8674(82)90085-x. [DOI] [PubMed] [Google Scholar]
- Little J. W. The SOS regulatory system: control of its state by the level of RecA protease. J Mol Biol. 1983 Jul 15;167(4):791–808. doi: 10.1016/s0022-2836(83)80111-9. [DOI] [PubMed] [Google Scholar]
- Lovett S. T., Clark A. J. Genetic analysis of regulation of the RecF pathway of recombination in Escherichia coli K-12. J Bacteriol. 1983 Mar;153(3):1471–1478. doi: 10.1128/jb.153.3.1471-1478.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madiraju M. V., Templin A., Clark A. J. Properties of a mutant recA-encoded protein reveal a possible role for Escherichia coli recF-encoded protein in genetic recombination. Proc Natl Acad Sci U S A. 1988 Sep;85(18):6592–6596. doi: 10.1073/pnas.85.18.6592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maenhaut-Michel G., Caillet-Fauquet P. Effect of umuC mutations on targeted and untargeted ultraviolet mutagenesis in bacteriophage lambda. J Mol Biol. 1984 Jul 25;177(1):181–187. doi: 10.1016/0022-2836(84)90064-0. [DOI] [PubMed] [Google Scholar]
- McCall J. O., Witkin E. M., Kogoma T., Roegner-Maniscalco V. Constitutive expression of the SOS response in recA718 mutants of Escherichia coli requires amplification of RecA718 protein. J Bacteriol. 1987 Feb;169(2):728–734. doi: 10.1128/jb.169.2.728-734.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McEntee K., Weinstock G. M. tif-1 mutation alters polynucleotide recognition by the recA protein of Escherichia coli. Proc Natl Acad Sci U S A. 1981 Oct;78(10):6061–6065. doi: 10.1073/pnas.78.10.6061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menzel R. A microtiter plate-based system for the semiautomated growth and assay of bacterial cells for beta-galactosidase activity. Anal Biochem. 1989 Aug 15;181(1):40–50. doi: 10.1016/0003-2697(89)90391-6. [DOI] [PubMed] [Google Scholar]
- Mieschendahl M., Griesser H. W., Müller-Hill B. lambda Immunity phase shift in a lambda N- -lacZ+ fusion. Mol Gen Genet. 1981;183(1):202–204. doi: 10.1007/BF00270164. [DOI] [PubMed] [Google Scholar]
- Morand P., Blanco M., Devoret R. Characterization of lexB mutations in Escherichia coli K-12. J Bacteriol. 1977 Aug;131(2):572–582. doi: 10.1128/jb.131.2.572-582.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nohmi T., Battista J. R., Dodson L. A., Walker G. C. RecA-mediated cleavage activates UmuD for mutagenesis: mechanistic relationship between transcriptional derepression and posttranslational activation. Proc Natl Acad Sci U S A. 1988 Mar;85(6):1816–1820. doi: 10.1073/pnas.85.6.1816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novick A., Weiner M. ENZYME INDUCTION AS AN ALL-OR-NONE PHENOMENON. Proc Natl Acad Sci U S A. 1957 Jul 15;43(7):553–566. doi: 10.1073/pnas.43.7.553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson K. R., Ossanna N., Thliveris A. T., Ennis D. G., Mount D. W. Derepression of specific genes promotes DNA repair and mutagenesis in Escherichia coli. J Bacteriol. 1988 Jan;170(1):1–4. doi: 10.1128/jb.170.1.1-4.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quillardet P., Devoret R. Damaged-site independent mutagenesis of phage lambda produced by inducible error-prone repair. Biochimie. 1982 Aug-Sep;64(8-9):789–796. doi: 10.1016/s0300-9084(82)80130-2. [DOI] [PubMed] [Google Scholar]
- Sancar A., Sancar G. B. DNA repair enzymes. Annu Rev Biochem. 1988;57:29–67. doi: 10.1146/annurev.bi.57.070188.000333. [DOI] [PubMed] [Google Scholar]
- Sancar A., Stachelek C., Konigsberg W., Rupp W. D. Sequences of the recA gene and protein. Proc Natl Acad Sci U S A. 1980 May;77(5):2611–2615. doi: 10.1073/pnas.77.5.2611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sassanfar M., Roberts J. W. Nature of the SOS-inducing signal in Escherichia coli. The involvement of DNA replication. J Mol Biol. 1990 Mar 5;212(1):79–96. doi: 10.1016/0022-2836(90)90306-7. [DOI] [PubMed] [Google Scholar]
- Sedgwick S. G., Goodwin P. A. Differences in mutagenic and recombinational DNA repair in enterobacteria. Proc Natl Acad Sci U S A. 1985 Jun;82(12):4172–4176. doi: 10.1073/pnas.82.12.4172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinagawa H., Iwasaki H., Kato T., Nakata A. RecA protein-dependent cleavage of UmuD protein and SOS mutagenesis. Proc Natl Acad Sci U S A. 1988 Mar;85(6):1806–1810. doi: 10.1073/pnas.85.6.1806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Story R. M., Weber I. T., Steitz T. A. The structure of the E. coli recA protein monomer and polymer. Nature. 1992 Jan 23;355(6358):318–325. doi: 10.1038/355318a0. [DOI] [PubMed] [Google Scholar]
- Sweasy J. B., Witkin E. M., Sinha N., Roegner-Maniscalco V. RecA protein of Escherichia coli has a third essential role in SOS mutator activity. J Bacteriol. 1990 Jun;172(6):3030–3036. doi: 10.1128/jb.172.6.3030-3036.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tessman E. S., Peterson P. Plaque color method for rapid isolation of novel recA mutants of Escherichia coli K-12: new classes of protease-constitutive recA mutants. J Bacteriol. 1985 Aug;163(2):677–687. doi: 10.1128/jb.163.2.677-687.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volkert M. R., Hartke M. A. Suppression of Escherichia coli recF mutations by recA-linked srfA mutations. J Bacteriol. 1984 Feb;157(2):498–506. doi: 10.1128/jb.157.2.498-506.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker G. C. Mutagenesis and inducible responses to deoxyribonucleic acid damage in Escherichia coli. Microbiol Rev. 1984 Mar;48(1):60–93. doi: 10.1128/mr.48.1.60-93.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W. B., Tessman E. S., Tessman I. Activation of protease-constitutive recA proteins of Escherichia coli by rRNA and tRNA. J Bacteriol. 1988 Oct;170(10):4823–4827. doi: 10.1128/jb.170.10.4823-4827.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willis D. K., Uhlin B. E., Amini K. S., Clark A. J. Physical mapping of the srl recA region of Escherichia coli: analysis of Tn10 generated insertions and deletions. Mol Gen Genet. 1981;183(3):497–504. doi: 10.1007/BF00268771. [DOI] [PubMed] [Google Scholar]
- Witkin E. M. Ultraviolet mutagenesis and inducible DNA repair in Escherichia coli. Bacteriol Rev. 1976 Dec;40(4):869–907. doi: 10.1128/br.40.4.869-907.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodgate R., Ennis D. G. Levels of chromosomally encoded Umu proteins and requirements for in vivo UmuD cleavage. Mol Gen Genet. 1991 Sep;229(1):10–16. doi: 10.1007/BF00264207. [DOI] [PubMed] [Google Scholar]
- Wu T. T. A model for three-point analysis of random general transduction. Genetics. 1966 Aug;54(2):405–410. doi: 10.1093/genetics/54.2.405. [DOI] [PMC free article] [PubMed] [Google Scholar]



