Abstract
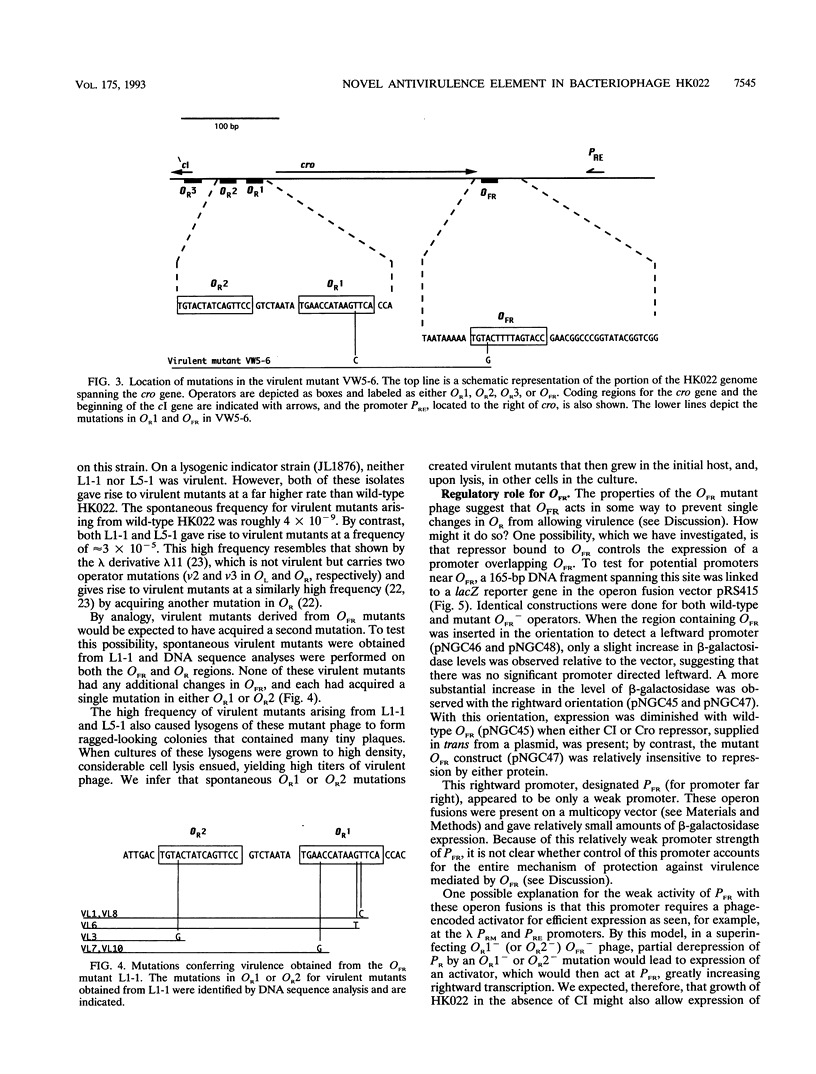
Lysogens of the temperate lambdoid phage HK022 are immune to superinfection by HK022. Superinfection immunity is conferred in part by the action of the HK022 CI repressor at the O.R operators. In this work, we have identified an additional regulatory element involved in immunity. This site, termed OFR (operator far right), is located just downstream of the cro gene, more than 250 nucleotides distant from OR. The behavior of phage containing a mutation in OFR suggests that the wild-type site functions as an antivirulence element. HK022 OFR- mutants were able to form turbid plaques indistinguishable from those of the wild type. However, they gave rise to virulent derivatives at a far higher frequency than the wild type (approximately 10(-5) for OFR- versus about 10(-9) for the wild type). This frequency was so high that cultures of HK022 OFR- lysogens were rapidly overgrown by virulent derivatives. Whereas virulent mutants arising from a wild-type OFR+ background contained mutations in both OR1 and OR2, virulent derivatives of the OFR- mutant phage contained a single mutation in either OR1 or OR2. We conclude that the wild-type OFR site functions to prevent single mutations in OR from conferring virulence. The mechanism by which OFR acts is not yet clear. Both CI and Cro bound to OFR and repressed a very weak rightward promoter (PFR). It is unlikely that repression of PFR by CI or Cro binding to OFR can account in full for the antivirulence phenotype conferred by this element, since PFR is such a weak promoter. Other models for the possible action of OFR are discussed.
Full text
PDF







Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Biere A. L., Citron M., Schuster H. Transcriptional control via translational repression by c4 antisense RNA of bacteriophages P1 and P7. Genes Dev. 1992 Dec;6(12A):2409–2416. doi: 10.1101/gad.6.12a.2409. [DOI] [PubMed] [Google Scholar]
- Borowiec J. A., Zhang L., Sasse-Dwight S., Gralla J. D. DNA supercoiling promotes formation of a bent repression loop in lac DNA. J Mol Biol. 1987 Jul 5;196(1):101–111. doi: 10.1016/0022-2836(87)90513-4. [DOI] [PubMed] [Google Scholar]
- Bushman F. D. The bacteriophage 434 right operator. Roles of O(R)1, O(R)2 and O(R)3. J Mol Biol. 1993 Mar 5;230(1):28–40. doi: 10.1006/jmbi.1993.1123. [DOI] [PubMed] [Google Scholar]
- Cam K. M., Oberto J., Weisberg R. A. The early promoters of bacteriophage HK022: contrasts and similarities to other lambdoid phages. J Bacteriol. 1991 Jan;173(2):734–740. doi: 10.1128/jb.173.2.734-740.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson N. G., Little J. W. Highly cooperative DNA binding by the coliphage HK022 repressor. J Mol Biol. 1993 Apr 20;230(4):1108–1130. doi: 10.1006/jmbi.1993.1229. [DOI] [PubMed] [Google Scholar]
- Choy H. E., Adhya S. Control of gal transcription through DNA looping: inhibition of the initial transcribing complex. Proc Natl Acad Sci U S A. 1992 Dec 1;89(23):11264–11268. doi: 10.1073/pnas.89.23.11264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Churchward G., Belin D., Nagamine Y. A pSC101-derived plasmid which shows no sequence homology to other commonly used cloning vectors. Gene. 1984 Nov;31(1-3):165–171. doi: 10.1016/0378-1119(84)90207-5. [DOI] [PubMed] [Google Scholar]
- Citron M., Schuster H. The c4 repressors of bacteriophages P1 and P7 are antisense RNAs. Cell. 1990 Aug 10;62(3):591–598. doi: 10.1016/0092-8674(90)90023-8. [DOI] [PubMed] [Google Scholar]
- Das A. How the phage lambda N gene product suppresses transcription termination: communication of RNA polymerase with regulatory proteins mediated by signals in nascent RNA. J Bacteriol. 1992 Nov;174(21):6711–6716. doi: 10.1128/jb.174.21.6711-6716.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deuschle U., Gentz R., Bujard H. lac Repressor blocks transcribing RNA polymerase and terminates transcription. Proc Natl Acad Sci U S A. 1986 Jun;83(12):4134–4137. doi: 10.1073/pnas.83.12.4134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhillon T. S., Dhillon E. K. Temperate coliphage HK022. Clear plaque mutants and preliminary vegetative map. Jpn J Microbiol. 1976 Oct;20(5):385–396. doi: 10.1111/j.1348-0421.1976.tb01004.x. [DOI] [PubMed] [Google Scholar]
- Flashman S. M. Mutational analysis of the operators of bacteriophage lambda. Mol Gen Genet. 1978 Oct 25;166(1):61–73. doi: 10.1007/BF00379730. [DOI] [PubMed] [Google Scholar]
- Flashner Y., Gralla J. D. Dual mechanism of repression at a distance in the lac operon. Proc Natl Acad Sci U S A. 1988 Dec;85(23):8968–8972. doi: 10.1073/pnas.85.23.8968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman D. I. Integration host factor: a protein for all reasons. Cell. 1988 Nov 18;55(4):545–554. doi: 10.1016/0092-8674(88)90213-9. [DOI] [PubMed] [Google Scholar]
- Goodman S. D., Nash H. A. Functional replacement of a protein-induced bend in a DNA recombination site. Nature. 1989 Sep 21;341(6239):251–254. doi: 10.1038/341251a0. [DOI] [PubMed] [Google Scholar]
- Grosschedl R., Schwarz E. Nucleotide sequence of the cro-cII-oop region of bacteriophage 434 DNA. Nucleic Acids Res. 1979 Mar;6(3):867–881. doi: 10.1093/nar/6.3.867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He B., Zalkin H. Repression of Escherichia coli purB is by a transcriptional roadblock mechanism. J Bacteriol. 1992 Nov;174(22):7121–7127. doi: 10.1128/jb.174.22.7121-7127.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochschild A. Detecting cooperative protein-DNA interactions and DNA loop formation by footprinting. Methods Enzymol. 1991;208:343–361. doi: 10.1016/0076-6879(91)08019-e. [DOI] [PubMed] [Google Scholar]
- JACOB F., WOLLMAN E. L. Etude génétique d'un bactériophage tempéré d'Escherichia coli. l. Le système genétique du bactériophage. Ann Inst Pasteur (Paris) 1954 Dec;87(6):653–673. [PubMed] [Google Scholar]
- Johnson A. D., Poteete A. R., Lauer G., Sauer R. T., Ackers G. K., Ptashne M. lambda Repressor and cro--components of an efficient molecular switch. Nature. 1981 Nov 19;294(5838):217–223. doi: 10.1038/294217a0. [DOI] [PubMed] [Google Scholar]
- Johnsrud L. Contacts between Escherichia coli RNA polymerase and a lac operon promoter. Proc Natl Acad Sci U S A. 1978 Nov;75(11):5314–5318. doi: 10.1073/pnas.75.11.5314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KAISER A. D., JACOB F. Recombination between related temperate bacteriophages and the genetic control of immunity and prophage localization. Virology. 1957 Dec;4(3):509–521. doi: 10.1016/0042-6822(57)90083-1. [DOI] [PubMed] [Google Scholar]
- Lin L. L., Little J. W. Autodigestion and RecA-dependent cleavage of Ind- mutant LexA proteins. J Mol Biol. 1989 Dec 5;210(3):439–452. doi: 10.1016/0022-2836(89)90121-6. [DOI] [PubMed] [Google Scholar]
- Little J. W., Hill S. A. Deletions within a hinge region of a specific DNA-binding protein. Proc Natl Acad Sci U S A. 1985 Apr;82(8):2301–2305. doi: 10.1073/pnas.82.8.2301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthews K. S. DNA looping. Microbiol Rev. 1992 Mar;56(1):123–136. doi: 10.1128/mr.56.1.123-136.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Oberto J., Clerget M., Ditto M., Cam K., Weisberg R. A. Antitermination of early transcription in phage HK022. Absence of a phage-encoded antitermination factor. J Mol Biol. 1993 Jan 20;229(2):368–381. doi: 10.1006/jmbi.1993.1040. [DOI] [PubMed] [Google Scholar]
- Oberto J., Weisberg R. A., Gottesman M. E. Structure and function of the nun gene and the immunity region of the lambdoid phage HK022. J Mol Biol. 1989 Jun 20;207(4):675–693. doi: 10.1016/0022-2836(89)90237-4. [DOI] [PubMed] [Google Scholar]
- Oehler S., Eismann E. R., Krämer H., Müller-Hill B. The three operators of the lac operon cooperate in repression. EMBO J. 1990 Apr;9(4):973–979. doi: 10.1002/j.1460-2075.1990.tb08199.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pirrotta V. Operators and promoters in the OR region of phage 434. Nucleic Acids Res. 1979 Apr;6(4):1495–1508. doi: 10.1093/nar/6.4.1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poteete A. R., Hehir K., Sauer R. T. Bacteriophage P22 Cro protein: sequence, purification, and properties. Biochemistry. 1986 Jan 14;25(1):251–256. doi: 10.1021/bi00349a035. [DOI] [PubMed] [Google Scholar]
- Poteete A. R., Ptashne M., Ballivet M., Eisen H. Operator sequences of bacteriophages P22 and 21. J Mol Biol. 1980 Feb 15;137(1):81–91. doi: 10.1016/0022-2836(80)90158-8. [DOI] [PubMed] [Google Scholar]
- Saha S., Lundqvist B., Haggård-Ljungquist E. Autoregulation of bacteriophage P2 repressor. EMBO J. 1987 Mar;6(3):809–814. doi: 10.1002/j.1460-2075.1987.tb04823.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schleif R. DNA looping. Annu Rev Biochem. 1992;61:199–223. doi: 10.1146/annurev.bi.61.070192.001215. [DOI] [PubMed] [Google Scholar]
- Simons R. W., Houman F., Kleckner N. Improved single and multicopy lac-based cloning vectors for protein and operon fusions. Gene. 1987;53(1):85–96. doi: 10.1016/0378-1119(87)90095-3. [DOI] [PubMed] [Google Scholar]
- Susskind M. M., Botstein D. Molecular genetics of bacteriophage P22. Microbiol Rev. 1978 Jun;42(2):385–413. doi: 10.1128/mr.42.2.385-413.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vologodskii A. V., Levene S. D., Klenin K. V., Frank-Kamenetskii M., Cozzarelli N. R. Conformational and thermodynamic properties of supercoiled DNA. J Mol Biol. 1992 Oct 20;227(4):1224–1243. doi: 10.1016/0022-2836(92)90533-p. [DOI] [PubMed] [Google Scholar]
- Wharton R. P., Brown E. L., Ptashne M. Substituting an alpha-helix switches the sequence-specific DNA interactions of a repressor. Cell. 1984 Sep;38(2):361–369. doi: 10.1016/0092-8674(84)90491-4. [DOI] [PubMed] [Google Scholar]
- Wu H. Y., Lau K., Liu L. F. Interlocking of plasmid DNAs due to Lac repressor-operator interaction. J Mol Biol. 1992 Dec 20;228(4):1104–1114. doi: 10.1016/0022-2836(92)90318-e. [DOI] [PubMed] [Google Scholar]




