Abstract
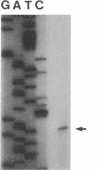
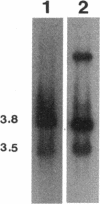
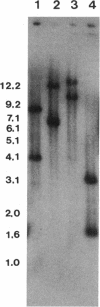
An electron transport regulatory gene, etrA, has been isolated and characterized from the obligate respiratory bacterium Shewanella putrefaciens MR-1. The deduced amino acid sequence of etrA (EtrA) shows a high degree of identity to both the Fnr of Escherichia coli (73.6%) and the analogous protein (ANR) of Pseudomonas aeruginosa (50.8%). The four active cysteine residues of Fnr are conserved in EtrA, and the amino acid sequence of the DNA-binding domains of the two proteins are identical. Further, S. putrefaciens etrA is able to complement an fnr mutant of E. coli. In contrast to fnr, there is no recognizable Fnr box upstream of the etrA sequence. Gene replacement etrA mutants of MR-1 were deficient in growth on nitrite, thiosulfate, sulfite, trimethylamine-N-oxide, dimethyl sulfoxide, Fe(III), and fumarate, suggesting that EtrA is involved in the regulation of the corresponding reductase genes. However, the mutants were all positive for reduction of and growth on nitrate and Mn(IV), indicating that EtrA is not involved in the regulation of these two systems. Southern blots of S. putrefaciens DNA with use of etrA as a probe revealed the expected etrA bands and a second set of hybridization signals whose genetic and functional properties remain to be determined.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Arnold R. G., DiChristina T. J., Hoffmann M. R. Inhibitor studies of dissimilative Fe(III) reduction by Pseudomonas sp. strain 200 ("Pseudomonas ferrireductans") Appl Environ Microbiol. 1986 Aug;52(2):281–289. doi: 10.1128/aem.52.2.281-289.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold R. G., Hoffmann M. R., Dichristina T. J., Picardal F. W. Regulation of Dissimilatory Fe(III) Reduction Activity in Shewanella putrefaciens. Appl Environ Microbiol. 1990 Sep;56(9):2811–2817. doi: 10.1128/aem.56.9.2811-2817.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batut J., Daveran-Mingot M. L., David M., Jacobs J., Garnerone A. M., Kahn D. fixK, a gene homologous with fnr and crp from Escherichia coli, regulates nitrogen fixation genes both positively and negatively in Rhizobium meliloti. EMBO J. 1989 Apr;8(4):1279–1286. doi: 10.1002/j.1460-2075.1989.tb03502.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colonna-Romano S., Arnold W., Schlüter A., Boistard P., Pühler A., Priefer U. B. An Fnr-like protein encoded in Rhizobium leguminosarum biovar viciae shows structural and functional homology to Rhizobium meliloti FixK. Mol Gen Genet. 1990 Aug;223(1):138–147. doi: 10.1007/BF00315806. [DOI] [PubMed] [Google Scholar]
- Devereux J., Haeberli P., Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 1):387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiChristina T. J. Effects of nitrate and nitrite on dissimilatory iron reduction by Shewanella putrefaciens 200. J Bacteriol. 1992 Mar;174(6):1891–1896. doi: 10.1128/jb.174.6.1891-1896.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eiglmeier K., Honoré N., Iuchi S., Lin E. C., Cole S. T. Molecular genetic analysis of FNR-dependent promoters. Mol Microbiol. 1989 Jul;3(7):869–878. doi: 10.1111/j.1365-2958.1989.tb00236.x. [DOI] [PubMed] [Google Scholar]
- Feng D. F., Doolittle R. F. Progressive sequence alignment as a prerequisite to correct phylogenetic trees. J Mol Evol. 1987;25(4):351–360. doi: 10.1007/BF02603120. [DOI] [PubMed] [Google Scholar]
- Galimand M., Gamper M., Zimmermann A., Haas D. Positive FNR-like control of anaerobic arginine degradation and nitrate respiration in Pseudomonas aeruginosa. J Bacteriol. 1991 Mar;173(5):1598–1606. doi: 10.1128/jb.173.5.1598-1606.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green J., Trageser M., Six S., Unden G., Guest J. R. Characterization of the FNR protein of Escherichia coli, an iron-binding transcriptional regulator. Proc Biol Sci. 1991 May 22;244(1310):137–144. doi: 10.1098/rspb.1991.0062. [DOI] [PubMed] [Google Scholar]
- Guest J. R. Oxygen-regulated gene expression in Escherichia coli. The 1992 Marjory Stephenson Prize Lecture. J Gen Microbiol. 1992 Nov;138(11):2253–2263. doi: 10.1099/00221287-138-11-2253. [DOI] [PubMed] [Google Scholar]
- Gunsalus R. P. Control of electron flow in Escherichia coli: coordinated transcription of respiratory pathway genes. J Bacteriol. 1992 Nov;174(22):7069–7074. doi: 10.1128/jb.174.22.7069-7074.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikemura T., Dahlberg J. E. Small ribonucleic acids of Escherichia coli. I. Characterization by polyacrylamide gel electrophoresis and fingerprint analysis. J Biol Chem. 1973 Jul 25;248(14):5024–5032. [PubMed] [Google Scholar]
- Iuchi S., Cameron D. C., Lin E. C. A second global regulator gene (arcB) mediating repression of enzymes in aerobic pathways of Escherichia coli. J Bacteriol. 1989 Feb;171(2):868–873. doi: 10.1128/jb.171.2.868-873.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iuchi S., Lin E. C. arcA (dye), a global regulatory gene in Escherichia coli mediating repression of enzymes in aerobic pathways. Proc Natl Acad Sci U S A. 1988 Mar;85(6):1888–1892. doi: 10.1073/pnas.85.6.1888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiley P. J., Reznikoff W. S. Fnr mutants that activate gene expression in the presence of oxygen. J Bacteriol. 1991 Jan;173(1):16–22. doi: 10.1128/jb.173.1.16-22.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisser S., Margalit H. Compilation of E. coli mRNA promoter sequences. Nucleic Acids Res. 1993 Apr 11;21(7):1507–1516. doi: 10.1093/nar/21.7.1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacInnes J. I., Kim J. E., Lian C. J., Soltes G. A. Actinobacillus pleuropneumoniae hlyX gene homology with the fnr gene of Escherichia coli. J Bacteriol. 1990 Aug;172(8):4587–4592. doi: 10.1128/jb.172.8.4587-4592.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers C. R., Nealson K. H. Bacterial manganese reduction and growth with manganese oxide as the sole electron acceptor. Science. 1988 Jun 3;240(4857):1319–1321. doi: 10.1126/science.240.4857.1319. [DOI] [PubMed] [Google Scholar]
- Myers C. R., Nealson K. H. Respiration-linked proton translocation coupled to anaerobic reduction of manganese(IV) and iron(III) in Shewanella putrefaciens MR-1. J Bacteriol. 1990 Nov;172(11):6232–6238. doi: 10.1128/jb.172.11.6232-6238.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obuekwe C. O., Westlake D. W., Cook F. D. Effect of nitrate on reduction of ferric iron by a bacterium isolated from crude oil. Can J Microbiol. 1981 Jul;27(7):692–697. doi: 10.1139/m81-107. [DOI] [PubMed] [Google Scholar]
- Perry K. A., Kostka J. E., Luther G. W., 3rd, Nealson K. H. Mediation of sulfur speciation by a black sea facultative anaerobe. Science. 1993 Feb 5;259(5096):801–803. doi: 10.1126/science.259.5096.801. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawers G., Böck A. Novel transcriptional control of the pyruvate formate-lyase gene: upstream regulatory sequences and multiple promoters regulate anaerobic expression. J Bacteriol. 1989 May;171(5):2485–2498. doi: 10.1128/jb.171.5.2485-2498.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawers G., Suppmann B. Anaerobic induction of pyruvate formate-lyase gene expression is mediated by the ArcA and FNR proteins. J Bacteriol. 1992 Jun;174(11):3474–3478. doi: 10.1128/jb.174.11.3474-3478.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawers R. G. Identification and molecular characterization of a transcriptional regulator from Pseudomonas aeruginosa PAO1 exhibiting structural and functional similarity to the FNR protein of Escherichia coli. Mol Microbiol. 1991 Jun;5(6):1469–1481. doi: 10.1111/j.1365-2958.1991.tb00793.x. [DOI] [PubMed] [Google Scholar]
- Spiro S., Guest J. R. FNR and its role in oxygen-regulated gene expression in Escherichia coli. FEMS Microbiol Rev. 1990 Aug;6(4):399–428. doi: 10.1111/j.1574-6968.1990.tb04109.x. [DOI] [PubMed] [Google Scholar]
- Spiro S., Guest J. R. Regulation and over-expression of the fnr gene of Escherichia coli. J Gen Microbiol. 1987 Dec;133(12):3279–3288. doi: 10.1099/00221287-133-12-3279. [DOI] [PubMed] [Google Scholar]
- Stewart V., Berg B. L. Influence of nar (nitrate reductase) genes on nitrate inhibition of formate-hydrogen lyase and fumarate reductase gene expression in Escherichia coli K-12. J Bacteriol. 1988 Oct;170(10):4437–4444. doi: 10.1128/jb.170.10.4437-4444.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strauch K. L., Lenk J. B., Gamble B. L., Miller C. G. Oxygen regulation in Salmonella typhimurium. J Bacteriol. 1985 Feb;161(2):673–680. doi: 10.1128/jb.161.2.673-680.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trageser M., Unden G. Role of cysteine residues and of metal ions in the regulatory functioning of FNR, the transcriptional regulator of anaerobic respiration in Escherichia coli. Mol Microbiol. 1989 May;3(5):593–599. doi: 10.1111/j.1365-2958.1989.tb00206.x. [DOI] [PubMed] [Google Scholar]
- Winkelman J. W., Clark D. P. Anaerobically induced genes of Escherichia coli. J Bacteriol. 1986 Jul;167(1):362–367. doi: 10.1128/jb.167.1.362-367.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann A., Reimmann C., Galimand M., Haas D. Anaerobic growth and cyanide synthesis of Pseudomonas aeruginosa depend on anr, a regulatory gene homologous with fnr of Escherichia coli. Mol Microbiol. 1991 Jun;5(6):1483–1490. doi: 10.1111/j.1365-2958.1991.tb00794.x. [DOI] [PubMed] [Google Scholar]