Abstract
Null-mutations of the Arabidopsis FKBP-like immunophilin TWISTED DWARF1 (TWD1) gene cause a pleiotropic phenotype characterized by reduction of cell elongation and disorientated growth of all plant organs. Heterologously expressed TWD1 does not exhibit cis-trans-peptidylprolyl isomerase (PPIase) activity and does not complement yeast FKBP12 mutants, suggesting that TWD1 acts indirectly via protein-protein interaction. Yeast two-hybrid protein interaction screens with TWD1 identified cDNA sequences that encode the C-terminal domain of Arabidopsis multidrugresistance-like ABC transporter AtPGP1. This interaction was verified in vitro. Mapping of protein interaction domains shows that AtPGP1 surprisingly binds to the N-terminus of TWD1 harboring the cis-trans peptidyl-prolyl isomerase-like domain and not to the tetratrico-peptide repeat domain, which has been shown to mediate protein-protein interaction. Unlike all other FKBPs, TWD1 is shown to be an integral membrane protein that colocalizes with its interacting partner AtPGP1 on the plasma membrane. TWD1 also interacts with AtPGP19 (AtMDR1), the closest homologue of AtPGP1. The single gene mutation twd1-1 and double atpgp1-1/atpgp19-1 (atmdr1-1) mutants exhibit similar phenotypes including epinastic growth, reduced inflorescence size, and reduced polar auxin transport, suggesting that a functional TWD1-AtPGP1/AtPGP19 complex is required for proper plant development.
INTRODUCTION
Parvulins, FK506-binding proteins (FKBPs) and cyclophilins represent three structurally unrelated classes of immunophilins known to function as cis-trans-peptidylprolyl isomerases (PPIases; Schiene and Fischer, 2000). The latter two are distinguished by their ability to bind different immunosuppressant drugs, either FK506/rapamycin or cyclosporin A (CsA). These products of soil-borne microorganisms are used to treat and prevent graft rejection in organ transplantation. Cyclophilin-CsA and FKBP12-FK506 complexes bind to calcineurin (PP2B), a Ca2+, calmodulin-regulated Ser/Thr-specific protein phosphatase, and thereby blocking Ca2+-dependent signaling (Cardenas et al., 1999; Harrar et al., 2001) leading to inhibition of T-cell activation. Additionally, CsA and FK506 play a role in reversing multidrug resistance (MDR) in several types of cancer by inhibiting the efflux of anticancer drugs (Cardenas et al., 1999).
Small FKBPs such as FKBP12 are thought to modulate signal transduction pathways (Harrar et al., 2001). FKBP12 functions as physiological regulator of the cell cycle. Cells from FKBP-deficient (FKBP12–/–) knock-out mice are arrested in G1 phase of the cell cycle (Aghdasi et al., 2001).
High-molecular-weight FKBPs are composed of one or more FKBP12-like domains and can be distinguished from their smaller counterparts by the presence of a tetratricopeptide repeat (TPR) domain (Das et al., 1998; Pratt et al., 2001) and a C-terminus that in most cases contains a putative calmodulin-binding domain (Harrar et al., 2001). Mammalian FKBP52, the best investigated example, is associated with hsp90 by its TPR domain in the native steroid hormone receptor complex (Silverstein et al., 1999) but plant high-molecular weight FKBPs bind plant hsp90 via the same TPR interaction as the mammalian homologues (Pratt et al., 2001; Kamphausen et al., 2002).
A recent proteomic investigation of Arabidopsis thylakoid lumen proteins describes 22 annotated FKBP-like proteins with predicted molecular weights from 12 to 72 kDa in the entire genome (Schubert et al., 2002). Although yeast seems to be viable without immunophilins (Dolinski et al., 1997), drastic phenotypes have been associated with mutations in individual plant immunophilins. Loss-of-function mutations in the cyclophilin40 homolog of Arabidopsis lead to reduction in number of juvenile leaves (Berardini et al., 2001). The Arabidopsis T-DNA mutant pasticcino1 (pas1), which lacks a 72-kDa FKBP is characterized by ectopic cell division, abnormally developed cotyledons and leaves, fusion of tissues, and impaired root development (Faure et al., 1998; Vittorioso et al., 1998). The Arabidopsis FKBP42 mutant twisted dwarf1 (twd1), results in a drastic reduction of cell elongation combined with a disoriented growth behavior (see Figure 1). Genetic analysis of twd1 null mutants demonstrates that TWD1 plays an important role in brassinosteroid reception or signal transduction (B. Schulz, B. Saal, D. Wanke, M. Lafos, H.Ü. Kolukisaoglu, B.P. Dilkers, and K.A.J. Feldman, unpublished results).
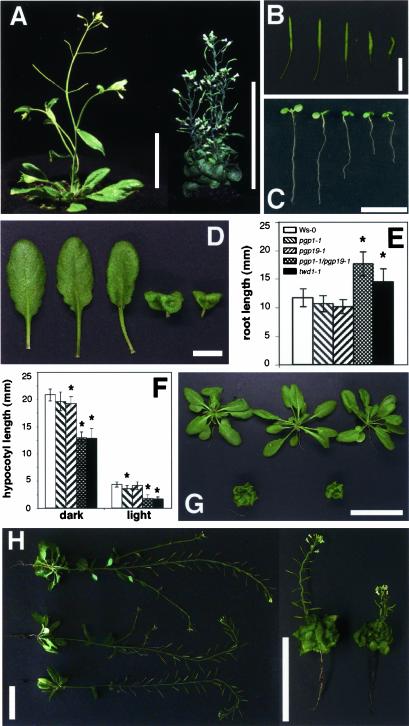
Figure 1.
The twisted dwarf1 (twd1) mutant displays a pleiotropic developmental phenotype resembling atpgp1-1/atpgp19-1 (atmdr1-1) double mutants. (A) Phenotype of soil-grown wild-type (left) and twd1-1 (right) plants at maturity. Bars, 5 cm. (B) Siliques of wild-type, atpgp1-1, atpgp19-1, atpgp1-1/atpgp19-1, and twd1-1 (from left to right) plants showing disoriented growth behavior. Bars, 1 cm. (C) Light grown seedlings 5 d after germination. From left to right: wild-type, atpgp1-1, atmdr1-1 (atpgp19-1), atmdr1-1/atpgp1-1, and twd1-1. Bar, 1 cm. (D) Rosette leaves of wild-type, atpgp1-1, atpgp19-1, atpgp1-1/atpgp19-1, and twd1-1 (from left to right) plants. Double mutant and twd1-1 show strongly reduced leaf expansion and strong epinastic growth behavior. Bar, 1 cm. (E) Dark-grown seedlings of atpgp1-1/atpgp19-1 (atmdr1-1), and twd1-1 plants have longer roots. Seedlings were grown on plate in darkness and root lengths. Root lengths were measured with a ruler (>10 seedlings) after 8 d and are presented as means plus SDs. Plant growth being statistically different (Mann-Whitney U test, p > 0.05) compared with wild-type control plants is indicated by an asterisks. (F) Seedlings of atpgp1-1/atpgp19-1 (atmdr1-1) and twd1-1 plants have longer hypocotyls. Seedlings were grown on plate in darkness or continuous white light. Hypocotyl lengths were measured with a ruler (> 10 seedlings) after 8 d and are presented as means plus SDs. Plant growth being statistically different (Mann-Whitney U test, p > 0.05) compared with wild-type control plants is indicated by an asterisks. (G) Phenotype of soil-grown plants after 40 d of culture. Top panel from left to right: wild-type, atpgp1-1, atpgp19-1 (atmdr1-1), bottom panel: atpgp1-1/atpgp19-1 (atmdr1-1) double mutant, twd1-1. Bar, 5 cm. (H) Reduced apical dominance in atpgp1-1/atpgp19-1 (atmdr1-1) and twd1-1 plants. Plants (left panel from top to bottom: wild-type, atpgp1-1, and atpgp19-1 (atmdr1-1); right panel from left to right: atpgp1-1/atpgp19-1 (atmdr1-1) and twd1-1) were cultured on soil for 70 d. Note the size bars differ between left and right panels.
We show here that TWD1 interacts with the MDR-like proteins AtPGP1 and AtPGP19, both members of the ABC transporter superfamily. AtPGP1 was the first MDR-like ABC transporter identified in Arabidopsis (Dudler and Hertig, 1992). Based on the Arabidopsis Genome Initiative sequence data (Arabidopsis Genome Initiative 2000), 22 members of the AtMDR subfamily have been annotated in the Arabidopsis genome (Sanchez-Fernandez et al., 2001; Martinoia et al., 2002). Like TWD1, AtPGP1 and AtPGP19 seem to be directly involved in plant growth processes. Downregulation of AtPGP1 by antisense inhibition causes a reduction of hypocotyl elongation in seedling grown under low light, whereas AtPGP1 overexpression leads to enhanced hypocotyl and root elongation (Sidler et al., 1998). Recently, Noh et al. (2001) and Murphy et al. (2002) have provided biochemical and genetic evidence suggesting that AtPGP1 together with its closest homologue AtMDR1, identified hereafter as AtPGP19 according to the nomenclature of Martinoia et al. (2002), are involved in polar auxin transport and auxinmediated development: auxin transport was greatly impaired in hypocotyls of atpgp19 and atpgp1 atpgp19 double mutants, and both proteins tightly bind the auxin transport inhibitor 1-naphthylphthalamic acid (NPA). atpgp1-1/atpgp19-1 (mdr1-1) double knock-out mutants exhibit epinastic cotyledons, shortened and curved hypocotyls in the dark, curled rosette leaves and dwarfed light-grown plants that strikingly resemble twd1 mutants.
FKBPs have been suggested to function as regulators of MDR-like ABC transporters (Cardenas et al., 1994), but any attempts to demonstrate a direct association with FKBP-like immunophilins have failed so far (Hemenway and Heitman, 1996; Mealey et al., 1999). Here we show, that TWD1 forms a protein-protein complex via the C-terminus of the ABC transporter AtPGP1 and that both colocalize and associate on the plasma membrane.
MATERIALS AND METHODS
Plant Growth Conditions
Seedlings were grown on 0.5× MS medium (Duchefa, Haarlem, The Netherlands) containing 1% sucrose under continuous light. Plants grown on soil were grown under white light (photon flux rate, 100 μmol m–2 s–1; 8-h light/16-h dark cycle at 20°C).
Yeast Two-hybrid Analysis
The coding region of the TWD1 gene from codon 1–337 was amplified by PCR (BUSUP: 5′ gga aaa acc atg gat gaa tct ctg gag cat caa act c, BUSdownB: 5′gga aaa agg atc ctt agc tct ttg act tag cac cac c) and cloned in frame via NcoI and BamHI restriction sites into pAS2, generating a protein fusion between TWD1 and the GAL4 DNA-binding domain (pAS2-BusB). The bait construct pAS2-BusB was used to screen an Arabidopsis cell suspension cDNA library inserted into pACT2 (Németh et al., 1998). Fast-growing colonies were selected on SD plates lacking leucine, tryptophan, and histidine with 50 mM 3-amino-1,2,4-triazole and β-gal–positive clones were sequenced.
To identify the interaction domain of the TWD1 protein, subclones of pAS2BusB were constructed. The PPIase-like domain (aa residues 1–163) and TPR domain omitting the membrane anchor (aa residues 163–337) of TWD1 were fused to the Gal4 BD of vector pAS2 (Clontech, Palo Alto, CA).
The nucleotide sequences encoding the C-termini of AtPGP10 (MIPS code At1g10680, base pairs 2812–3681), AtPGP13 (At1g27940, base pairs 2872–3735), AtPGP14 (At1g28019, base pairs 2876–3744), and AtPGP19 (At3g28860, base pairs 2893–3756) were cloned by two-step RT-PCR. Therefore, total RNA from Arabidopsis thaliana (Wassilewskija ecotype) grown in liquid culture under mixotrophic conditions was prepared using the RNA Plant Mini Kit (Qiagen, Hilden, Germany). cDNA was generated from 1 μg of RNA using the M-MLV Reverse Transcriptase, RNase H– Point Mutant DNA Polymerase (Promega, Madison, WI) and the following gene-specific primers located in the 3′ untranslated region of the genes: AtPGP10: 5′ ttc ctt tca aga atg aat agc; AtPGP13: 5′ gtg tcc aga tat tcc tga cac; AtPGP14: 5′ tag ata ttc cca aca caa tcg; and AtPGP19: 5′ cat agt tca gtc tta tgt tcc. The C-termini were inserted into pACT2 after PCR amplification using Vent DNA Polymerase (New England Biolabs, Frankfurt, Germany), and the following primers (UP/LP): AtPGP10: 5′ acg gaa ttc tgg gtg aag tgt tgg ctc tag/5′ acg ctc gag tta agg atg atg gcg ctg ccg; AtPGP13: 5′ acg gaa ttc tgt cgg aaa cgc ttg ctt tga/5′ acg ctc gag tca cag tac ttc ttg aag act c; AtPGP14: 5′ acg gaa ttc tgg cgg aaa cgc ttg cgt taa cc/5′ acg ctc gag tca cac cgc ttc ttg aag act c; and AtPGP19: 5′ acg cca tgg aaa ctc tca gtc ttg ctc ctg/5′ acg gga tcc tca aat cct atg tgt ttg aag c. AtPGP2 (At4f25860) was amplified by PCR from the plasmid Y97 using the primers (UP/LP): 5′ acg gaa ttc tgg aga cat tgg ctc tag ctc cg/5′ acg ctc gag tta agg ttg ttg ctg ctg ctg. All RT-PCR products were sequenced to verify the absence of mistakes.
For interaction analysis, three to five independent transformants of two independent constructs were tested for HIS auxotrophy and LacZ (β-galactosidase) reporter activity. Single colonies were resuspended in 1 ml of sterile water and 5 μl each were spotted on SD plates lacking leucine, tryptophan, and histidine containing 25 mM 3-amino-1,2,4-triazole. Another 5 μl were spotted on plates with selective media supplemented with 30 μg/ml 5-bromo-4-chloro-3-indolyl-α-d-galactopyranoside (X-α-Gal). Growth was judged after 3 d. β-galactosidase activity was quantified by liquid culture assay using standard protocols.
Recombinant Expression of TWD1- and AtPGP1 Protein
PCR-amplified TWD1 (Primers: JOE1: 5′ cgg gat ccc agg ttg att tgg gaa ata atg g and 6118: 5′ ggg ggt aga tct ttc acg ttg) was restricted with BamHI and BglII and inserted into the BamHI-site of pQE31 (Qiagen). Restriction of the PCR product with BamHI and SspI removed the putative membrane anchor and the resulting fragment corresponding to aa residue 1–324 was ligated into BamHI- and SmaI-digested pQE31 (pTWD1–3). TWD1–3 peptides as N-terminal RGSH6 tagged fusions were purified on Ni-NTA agarose (Qiagen) under native conditions and dialyzed twice against 50 mM MOPS, pH 7.0. Immunodetection of TWD1–3 on Western blots was performed with polyclonal antiserum against TWD1-1 peptide (see Immunocytochemistry and Confocal Fluorescence Microscopy Analysis).
The insert of clone pACT2–4F12 encoding the C-terminus of AtPGP1 was cut out from the two-hybrid vector using BglII sites flanking the insert and ligated in-frame into the BamHI site of pQE32 (Qiagen). The resulting peptides were expressed as an N-terminal 6× His-tagged fusion protein in Escherichia coli strain BL21D3 pLysC (Stratagene, La Jolla, CA) and immunoprobed with polyclonal antiserum against AtPGP1 peptide (Sidler et al., 1998).
In Vitro Binding Assays
Ni-NTA-affinity–purified TWD1–3 peptides were immobilized to affigel-10 beads (Bio-Rad, Hercules, CA) as recommended by the manufacturer. Matrix-bound TWD1–3 (1 μg) was incubated for 1 h at 4°C with cleared E. coli supernatants from cells overproducing the C-terminus of AtPGP1, H+-ATPase AHA2 (expressed from plasmid pMP900; Fuglsang et al., 1999) or vector control lysates diluted twice with binding buffer (50 mM MOPS, pH 7.4, 100 mM NaCl, 10% [vol/vol] glycerol, 2 mM CaCl2, 2 mM MgCl2). After washing of the matrix-protein complex with binding buffer, the bound proteins were eluted by boiling the matrix with probe buffer and equal volumes of bound and nonbound protein were detected by Western blot analysis using monoclonal anti-RGSH6, antipenta His (both from Qiagen) and anti-ACA4N27 (Geisler et al., 2000) recognizing the AtPGP1 C-terminus, TWD1–3, and the GST fused to the AHA2 C-terminus, respectively. Individual bands were quantified using the Scion Image software 1.63 (http://www.scioncorp.com).
Membrane Fractionation
Equal volumes of Arabidopsis microsomes, separated by continuous sucrose gradient centrifugation, were blotted onto nitrocellulose membranes and probed with anti-AHA3 antiserum (no. 762; 1:3000), anti-V-ATPase antiserum (2E7; 1:200), anti-BIP antiserum (tobacco BIP; 1:5000), anti-AtPGP1 antiserum (1:1000), anti-TWD1 antiserum (1:1000), and monoclonal antibodies anti-HA (clone 12CA5; 1:3000, Roche Diagnostics, Basel, Switzerland) and anti c-myc (clone 9E10; 1:3000, Roche Diagnostics) as described in Geisler et al. (2000).
Plasma membranes were purified by one-step aqueous two-phase partitioning of Arabidopsis microsomes in a 6.2% (wt/wt) dextran T500/PEG4000 phase system containing 3 mM KCl and 5 mM potassium phosphate buffer, pH 7.8. Arabidopsis microsomes were prepared from 75 g of a 4-d-old cell suspension culture (cell line T87) grown in the dark (Axelos et al., 1992).
Transgenic Plants
Arabidopsis plants, ecotype Columbia, were transformed with an expression construct for a hemagglutinin (HA)-tagged TWD1 protein. Therefore, the entire open reading frame of TWD1 was amplified by PCR (TAGfor1: 5′ gac ctc gag gtt aac aat ggc tta and TAGrev1: 5′ cgc gga tcc gga gcg taa tca ggt aca tcg) and inserted into the BamHI site of cloning vector pRTΩ-NotI (Überlacker and Werr, 1996). To fuse the 9-aa-long HA1-tag to the TWD1 peptide, the construct was digested with XhoI and BamHI to remove the Ω-sequence from tobacco mosaic virus, which was replaced by an HA1-tag with compatible ends (pRTΩ-NotI3/4T). The cassette containing the CaMV 35S promoter and the HA-tagged TWD1 gene was excised with AscI and inserted after Klenow fill-in into the blunted HindIII site of binary vector pPTV-BAR (B. Schulz, B. Saal, D. Wanke, M. Lafos, H.Ü. Kolukisaoglu, B.P. Dilkes, and K.A.J. Feldman, unpublished results). The resulting binary construct pPTV3/4/2T was used to transform Arabidopsis via vacuum infiltration using Agrobacterium strain GV3101. BASTA resistant transformants were selected on soil and a line containing a single copy T-DNA was selected by Southern blot hybridization (our unpublished results).
Immunocytochemistry and Confocal Fluorescence Microscopy Analysis
For TWD1 antiserum production, a partial peptide (TWD1-1) comprising the first 187 aa of TWD1 was cloned into pET3-His, expressed as a 6× His-tag version in E. coli strain Bl21DE3 (Stratagene) and purified under denaturing conditions by Ni-NTA agarose chromatography. The purified protein was subjected to preparative SDS-PAGE and the eluted band was used for anti-serum production performed by BioGenes Inc. (Berlin, Germany) using standard protocols.
Protoplasts from leaves of HA-TWD1–expressing plants were prepared and fixed, and immunocytochemistry was performed as described in Geisler et al., 2000. Incubations with monoclonal anti-HA high affinity antibody (clone 12CA5; Roche, Rotkreuz, Switzerland) and secondary anti-mouse antibody coupled to FITC (Jackson ImmunoResearch Laboratories, West Grove, PA) were performed for 1 h with a 1:100 dilution. FITC and TRITC fluorescence was detected with the corresponding filter sets and stored images were colored as green (FITC) or false colors (TRITC) using Adobe PhotoShop 5.5 (Adobe Systems Inc., San Jose, CA).
Coimmunoprecipitation
Immunoprecipitations were carried out using the Seize X Protein G Immunoprecipitation Kit and the imidoester cross-linker DTBP according to the manufacturer (Pierce, Rockford, IL). Approximately 500 μg of Arabidopsis microsomes, prepared as described above, were cross-linked at 4°C and membrane proteins were solubilized using 2% (vol/vol) TX-100. Cleared lysates were loaded on anti-AtPGP1 columns made by binding and crosslinking of the antiserum to protein G. After washing, bound proteins were eluted, separated by PAGE in the presence or absence of DTT, and probed against anti-TWD1 antisera.
TWD1 Affinity Chromatography
Solubilized microsomal proteins were prepared from 6 d, light-grown, HATWD1–overexpressing seedlings and separated by anion exchange chromatography as described previously (Murphy et al., 2002) with the exception that the phase-separation enrichment of plasma membrane proteins and preliminary gel permeation chromatography steps were eliminated and the solubilization buffer contained 50 μM naphthylphthalamic acid (NPA) where noted. After SDS-PAGE, Western blots were prepared utilizing anti-HA-epitope (Sigma, St. Louis, MO) and alkaline phosphatase–conjugated goat anti-rabbit polyclonal antibodies and visualized with Lumiphos (Roche, Indianapolis, IN) reagent.
Separately, native HA-tagged TWD1 was purified from microsomal membrane proteins of HA-TWD1–overexpressing plants solubilized with 50 μM NPA (see above) utilizing immobilized anti-HA affinity resin (Roche). After extensive washing with PBS, solubilized microsomal proteins were incubated with the affinity matrix for 4 h at 4°C, and washed extensively with PBS. Immobilized proteins were then eluted with 30 μM NPA in PBS and visualized by SDS PAGE and Western blotting with a polyclonal anti-AtPGP1 antibody (Sidler et al., 1998) and goat anti-rabbit antibody as above.
Auxin Transport Assays
Auxin transport assays were conducted on intact light grown seedlings as described previously (Murphy et al., 2000; Noh et al., 2001), with the following exceptions: seedlings (WS wild-type, twd1-1, atpgp1-1, atpgp19-1, and atpgp1-1/atpgp19-1) used in the transport assays were grown in light on 1% phytagar plates containing 0.25× MS (pH 5.2) and 1% sucrose until hypocotyl lengths reached 5 mm. Before assay, 10 seedlings were transferred to vertically discontinuous filter paper strips saturated in 0.25× MS and allowed to equilibrate for 1.5 h. Auxin solutions used to measure transport were made up in 0.25% agarose containing 2% DMSO and 25 mM MES (pH 5.2). Using microscope-guided micromanipulators, a 0.1-μl microdroplet containing 500 nM unlabeled IAA and 500 nM [3H]IAA (specific activity 25 Ci/mmol, American Radiochemical, St. Louis, MO) was placed on the apical tip of seedlings. Seedlings were then incubated in the dark for 5 h. After incubation, the upper hypocotyls and cotyledons were removed, and a 2-mm section centered on the root-shoot transition zones was harvested, along with a 4-mm basal section of each root.
RESULTS
Loss-of-function mutation of the Arabidopsis FKBP-like immunophilin TWISTED DWARF1 (TWD1) gene results in dramatic differences in growth and organ development in comparison to wild type. The pleiotropic mutant phenotype is characterized by reduction of cell elongation and disorientated growth of nearly all plant organs. Leaves and cotyledons of twd1-1 show epinastic growth, hypocotyls are shorter and root growth is reduced in the light, but enhanced in the dark. Cell elongation in twd1-1 plants is severely impaired, which results in a dwarf phenotype (see Figure 1; Schulz et al., unpublished results).
Isolation of AtPGP1 as a TWD1-interacting Protein
Heterologously expressed TWD1 does not exhibit a PPIase activity (Kamphausen et al., 2002) and does not complement yeast FKBP12 shown by its inability to restore the sensitivity toward rapamycin, which is caused by disruption of the FKBP12 gene in yeast (our unpublished results). Therefore, we assumed that TWD1 acts indirectly via protein-protein interactions.
Screening of an Arabidopsis cDNA library made from suspension culture with the entire cytosolic domain of TWD1 as bait (BusB, TWD1 amino acid [aa] residues 1–337, Figure 2B) resulted in more than 1800 His-auxotrophic clones. Fortyeight β-galactosidase-positive prey clones were sequenced and six of those encoded C-terminal peptides of multidrug resistance-like ABC transporter (ABCB1) AtPGP1 (Dudler and Hertig, 1992; Martinoia et al., 2002). Colony hybridization with these cDNA clones revealed that ∼7% of all clones harbored AtPGP1-like sequences. The specificity of TWD1 interaction with AtPGP1 was confirmed using unrelated CBL1 and CIPK proteins (Shi et al., 1999) as positive and GAL4-binding domain (BD) or activation domain (AD) alone not interacting with TWD1 or AtPGP1 as negative controls, respectively (Figure 2A).
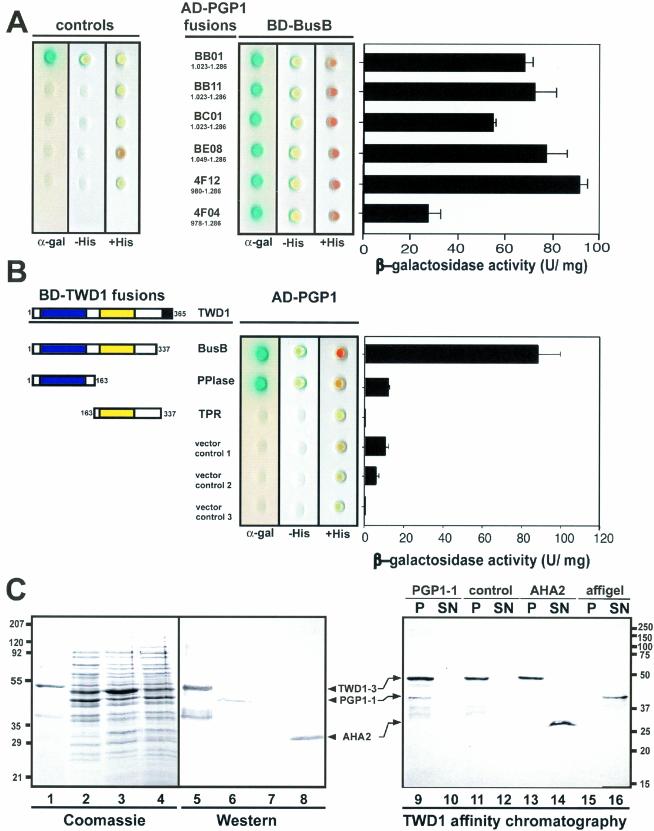
Figure 2.
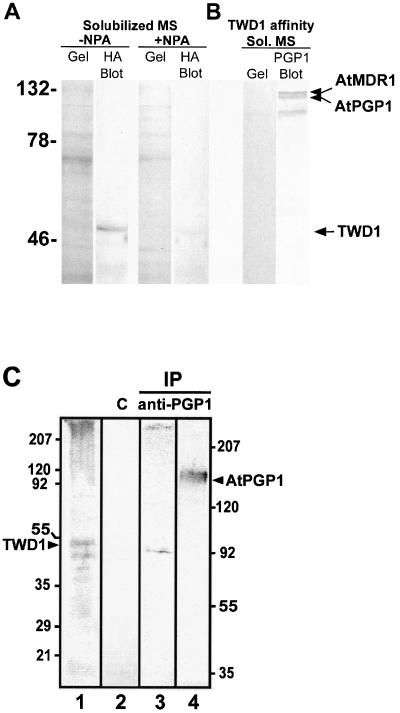
Analysis of TWD1-AtPGP1 interaction. (A) Interaction between TWD1 (BD-BusB) and six AtPGP1 clones fused to the GAL4 activation domain (AD) isolated in a yeast two-hybrid screen. Controls are from top to bottom: pGBT9.BS-CBL1/pGAD-CIPK (positive control), pGBT9.BS vector/pGAD vector, BD-BusB/AD vector, BD vector/AD-4F12, and AD vector/BD vector (negative controls). (B) The PPIase-like domain of TWD1 is responsible for the interaction with AtPGP1. The PPIase-like and the TPR-domain of TWD1 as GAL4 binding domain (BD) fusions were tested against activation domain AD-PGP1 fusion (clone 4F12). Colored boxes represent the following putative functional domains: blue, cis-trans-peptidyl proly isomerase domain; yellow, tetratrico-peptide repeat; black, membrane anchor. Transformants were analyzed for histidine auxotrophy and LacZ (β-galactosidase) reporter activity. Single colonies were spotted on selective media plates supplemented with X-α-Gal. LacZ reporter activities were quantified by liquid culture assays and are displayed as units per mg protein; error bars represent SDs from three to five independent transformants. (C) Ni-affinity–purified TWD1–3 (lane 1 and 5) and cleared total E. coli lysates containing the expressed C-termini of AtPGP1 (lane 2 and 6), the vector control (lane 3 and 7) and the C-terminus of Arabidopsis H+-ATPase AHA2 (lane 4 and 8) were visualized as Coomassie Blue stain (left panel) and immunoprobed (middle panel) as described in MATERIALS AND METHODS. A TWD1 affinity matrix was incubated with cleared E. coli lysates containing the expressed C-termini of AtPGP1, the vector control or the C-terminus of Arabidopsis H+-ATPase AHA2. As negative control, empty affigel beads were incubated with the AtPGP1-1 lysate. Equal volumes of matrix-eluted (P) as well as unbound proteins (SN) were separated by PAGE, and immunoprobed using the antisera described above (see MATERIALS AND METHODS).
All TWD1-interacting AtPGP1 prey constructs coded for the C-terminus of AtPGP1 carrying the C-terminal nucleotide binding fold covering the Walker A and B boxes and the intermediate ABC signature (Rea et al., 1998; Martinoia et al., 2002). Similar galactosidase activities with all AtPGP1 clones suggest that a peptide of 237 aa (aa residues 1049–1286) is sufficient for interaction.
Interaction with AtPGP1 Is Mediated by the PPIase-like Domain of TWD1
To assess whether the TPR domain—a 34-aa long protein-protein interaction motif (Owens-Grillo et al., 1996; Das et al., 1998; Pratt et al., 2001) localized in the C-terminal part of TWD1—was responsible for the interaction with AtPGP1, we generated GAL4-BD fusions covering the PPIase-like (aa residues 1–163) and the TPR domains (aa residues 163–337) of TWD1. Surprisingly, AtPGP1 interacted only with the N-terminus containing the PPIase-like domain, but not with the TPR domain containing part of TWD1 (Figure 2B), as can be judged from the β-galactosidase activity test on colonies as well as growth on plates lacking histidine.
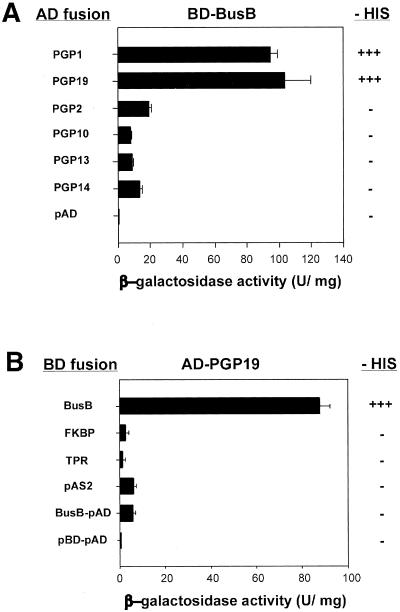
Very recently, both AtPGP1 and its closest homolog At-PGP19 (AtMDR1) were copurified by NPA affinity chromatography (Murphy et al., 2002) and have been implicated in polar auxin transport (Noh et al., 2001; Luschnig 2002). To test whether the C-terminus of AtPGP19 was also able to bind to TWD1, we generated GAL4-AD fusions of a homologous stretch of AtPGP19 (aa residues 965-1252). AtPGP19 interacted specifically with TWD1 in the yeast two-hybrid system, whereas the C-termini of related multidrug-resistance ABC transporters AtPGP2, AtPGP10, AtPGP13, and AtPGP14 did not (Figure 3A). β-galactosidase and HIS-auxotrophy assays suggest similar strengths of interaction for AtPGP1 and AtPGP19 with the TWD1 construct BusB. However, the interacting domain of AtPGP19 could not be mapped clearly to either the PPIase-like or TPR domain of TWD1 (Figure 3B).
Figure 3.
The entire TWD1 protein is essential for interaction with AtMRP19. (A) TWD1 interacts specifically with AtPGP1 and AtPGP19 TWD1 fused to a GAL4 binding domain (BD-Bus) was tested for interaction with AD fusion of AtPGPs that are closely related to AtPGP1. See MATERIALS AND METHODS for accession numbers. (B) Mapping of TWD1 domains that interact with AtPGP19. TWD1 fragments fused to a GAL4 binding domain (BD) tested for interaction with AD-AtPGP19 are represented by boxes and correspond to Figure 2. Activation of histidine growth reporter (growth on–HIS) is indicated by + and –; LacZ reporter activities are displayed as units per mg protein. Error bars represent SDs from three to five independent transformants.
In Vitro Protein Interaction Assay
To verify the two-hybrid data in vitro, AtPGP1 peptide 4F12 was expressed in E. coli, which was shown as Coomassie stain and Western detection using anti-RGSH6 in Figure 2C (lanes 2 and 6). TWD1–3 (aa residue 1–337) was affinity-purified on Ni-NTA agarose (Figure 2C, lane 1, Coomassie stain, and lane 5, Western detection using anti-penta His) and immobilized on affigel beads. The TWD1 affinity matrix was able to quantitatively sediment the AtPGP1 C-terminus of 42 kDa from soluble E. coli extracts shown by Western analysis of corresponding amounts of bound (P) and unbound fractions (SN). Monoclonal anti-RGSH6 and anti-penta His were used to recognize the AtPGP1 C-terminus and TWD1–3, respectively (Figure 2C, lanes 9 and 10). This high ratio (100%) indicates the specificity of TWD1-AtPGP1 interaction. As eukaryotic glycoproteins are not glycosylated when expressed in E. coli, this result suggests that the interaction of TWD1 with AtPGP1 is dependent on primary amino acid sequence interactions rather than interactions of TWD1 with carbohydrate moieties.
Using the same pair of antisera, no AtPGP1 protein was detected in bound fractions of controls in which a vector control lysate (Figure 2C, lane 11) or the empty affigel resin (Figure 2B, lane 15) was used. As a specific control, we tested the C-terminus of plasma membrane H+-ATPase AHA2, which binds to 14-3-3 proteins (Fuglsang et al., 1999). The AHA2 C-terminus (aa residues 850–948) that was expressed as GST fusion of ∼30 kDa and immunodetected with anti-ACA4N27 antiserum (Geisler et al., 2000) against the GST-tag did not bind to the TWD1 matrix (Figure 2C, lane 13).
TWD1 and AtPGP1 Form a Complex on the Plasma Membrane
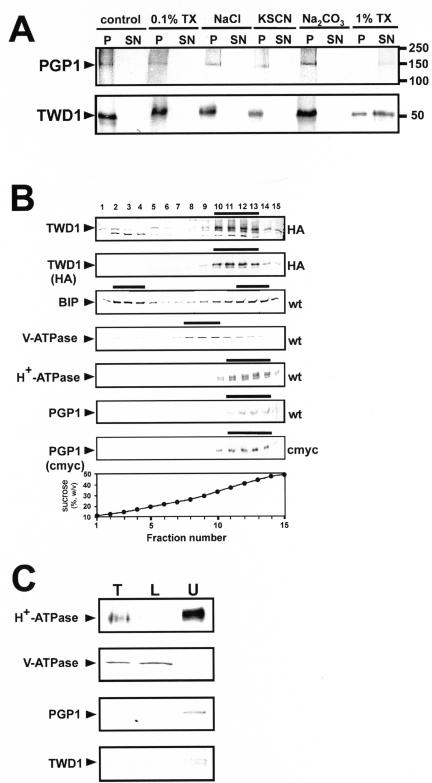
The C-terminus of TWD1 contains a hydrophobic α-helical region (residues 339–357) with the potential to form a membrane anchor predicted by hydrophobicity analysis. To demonstrate that TWD1 is indeed a membrane-anchored protein, the TWD1 protein was N-terminally tagged with a HA-epitope and expressed in transgenic plants. Pellets of microsomal membrane fractions prepared from HA-TWD1–expressing plants were treated either with chaotropic agent KSCN, high salt, carbonate, or TX-100. TWD1 could only be released from microsomes by solubilization with high concentrations (1% vol/vol) of the detergent TX-100 (Figure 3A), indicating that TWD1 is in fact a membrane anchored, rather than peripheral membrane protein.
AtPGP1 has been localized in the plasma membrane (Sidler et al., 1998), suggesting that TWD1 resides as well on the plasma membrane. To test this assumption, membranes prepared from Arabidopsis plants overexpressing a HA-epitope–tagged form of TWD1 were separated by linear sucrose gradient density centrifugation. Both polyclonal anti-TWD1 antiserum and a monoclonal anti-HA antibody detected HA-TWD1 (48 kDa) in fractions 10–13 (sucrose concentrations of 34 and 44%) of the sucrose gradient (Figure 4B). TWD1 colocalized with the plasma membrane marker H+-ATPase AHA3 and AtPGP1 (same distribution of peak fractions 11–14, Figure 4B). Markers for other membranes, such as the vacuolar V-ATPase subunit B or BIP, an ER-specific marker, cross-reacted with other fractions. Anti-TWD1 recognized additionally a smaller 40-kDa protein; detection of this protein with the monoclonal anti-HA antibody suggested that it represents a degradation product of TWD1.
Figure 4.
TWD1 is a plasma membrane-anchored protein. (A) Microsomal fractions expressing an HA-epitope–tagged version of TWD1 or a c-Myc epitope–tagged version of AtPGP1 were treated with 0.1% TX-100, 2 M NaCl, 1 M KSCN, 200 mM Na2CO3, or 1% TX-100. Membranes were pelleted, and supernatants were precipitated with TCA and subjected to PAGE. TWD1 and AtPGP1 were detected using anti-HA and antic-Myc antibodies. (B) Microsomes of wild-type (wt) and transgenic Arabidopsis plants ectopically expressing c-Myc- or HA-epitope–tagged AtPGP1 (c-Myc) and TWD1 (HA), respectively, were subjected to linear sucrose density gradient fractionation. Fractions were immunoprobed against given marker enzymes as described in Geisler et al. (2000). Transgenic plant material was probed additionally against antic-Myc (c-Myc) and anti-HA (HA), respectively. Immunopositive peak fractions are highlighted by bars. (C) Microsomal fractions obtained from aqueous two-phase partitioning of Arabidopsis suspension culture were probed with anti-TWD1 and anti-AtPGP1 antisera. Efficient partitioning of total microsomes (T, 10 μg of protein) to the lower phase (L, 10 μg of protein) or to the upper phase (U, 5 μg of protein) was ascertained by Western blot analysis using antisera against the marker proteins vacuolar V-ATPase and the plasma membrane–bound H+-ATPase.
To confirm these data, microsomal membranes from wild-type Arabidopsis suspension cultures were separated by aqueous two-phase partitioning. Efficient partitioning of internal cell membranes to the bottom phase and plasma membranes to the top phase was ascertained by Western blotting using anti-sera against the vacuolar V-ATPase, plasma membrane–bound AtPGP1, and H+-ATPase (Geisler et al., 2000), revealing no cross-contamination of both membrane types (Figure 4C). A protein band corresponding to the expected size of TWD1 was detected in the top fraction of phase partitioning with anti-TWD1 antisera confirming its plasma membrane location (Figure 4C).
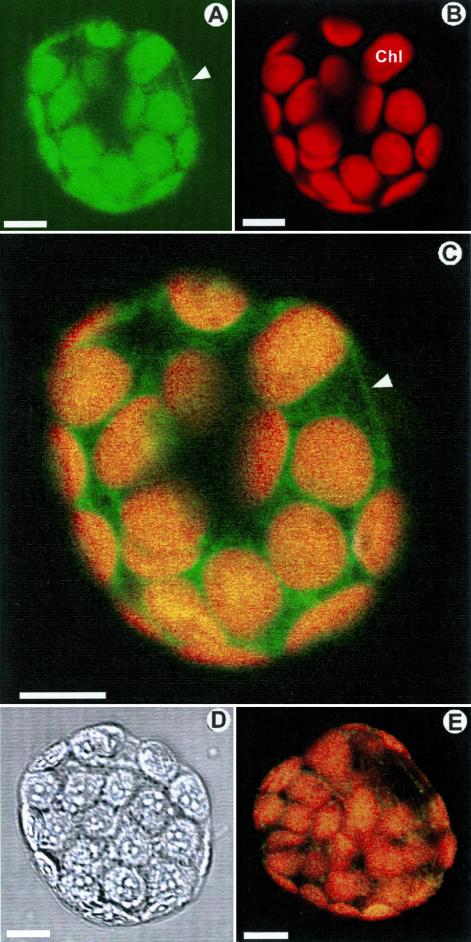
These biochemical fractionation data were supported by cellular immunolocalization of HA-TWD1 protein in transgenic plant cells by laser scanning microscopy. Protoplasts from leaves were treated with anti-HA antibody, which was decorated with a FITC-conjugated secondary antiserum. Optical sections showed that transgenic protoplasts were FITC labeled at the periphery (green in Figure 5, A and C), which is consistent with a plasma membrane localization of TWD1. Protoplasts treated only with the secondary antiserum revealed no background and wild-type material treated with both antibodies resulted in only very faint peripheral background (Figure 5E).
Figure 5.
Immunolocalization of TWD1 in the plasma membrane. (A) Fluorescence of a protoplast from HA-TWD1 plants immunoprobed with anti-HA antibody recorded with FITC filter settings. (B) The same protoplast sample detected with TRITC filter settings. (C) Superimposing of A and B. (D) The same protoplast as in A and B in bright field illumination. (E) Superimposing of fluorescence of wild-type protoplast treated as described A using FITC and TRITC filter settings. Images A–C, and E represent internal optical sections generated by laser scanning confocal microscopy. Note that green fluorescence in the chloroplasts (Chl) is not due to FITC fluorescence but to green coloring of chlorophyll autofluorescence of chloroplasts. Arrowheads mark the fluorescence of the plasma membrane. Bars, 10 μm.
Subsequent recording of chlorophyll autofluorescence using TRITC filter settings showed that this fluorescence is limited exclusively to the chloroplasts (red in Figure 5B), which revealed no peripheral fluorescence around the protoplasts. Superimposed false green and red images represent images obtained with FITC and TRITC filters.
A different strategy to detect TWD1-AtPGP interaction in plant cells was followed by showing that on the one hand TWD1 is excluded from solubilized microsomal protein preparations separated by anion exchange chromatography after treatments with NPA (Figure 6A). On the other hand, using HA-TWD1 protein, isolated from overexpressing plants as a ligand to anti-HA-epitope resin, we were able to immobilize AtPGP proteins from microsomal membrane preparations and visualize them using a polyclonal anti-PGP1 antibody (Sidler et al., 1998). NPA treatments elute AtPGP1 and AtPGP19, illustrating a specific interaction between TWD1 and those two transporters (Figure 6B). Identities of both upper bands was verified by MALDI analysis of tryptic fragments (unpublished results), whereas the identity of a third band of lower molecular weight is unknown.
Figure 6.
In vivo interaction between TWD1 and AtPGPs. (A) TWD1 is excluded from microsomal fractions by naphtylphtalamic acid (NPA). Microsomal proteins were solubilized in the presence and absence of NPA, fractionated by anion exchange chromatography and separated on SDS-PAGE gels. Western blots of these gels were probed with a monoclonal HA antibody (Sigma, St. Louis, MO). (B) Copurification of TWD1 and AtPGPs via TWD1 affinity chromatography. HA-TWD1 protein, purified from overexpressing plants in the presence of NPA, was bound to anti-HA affinity resin. Solubilized microsomal proteins were incubated with this matrix and proteins were eluted after washing with PBS with 30 μM NPA. Eluted proteins were separated on SDS-PAGE gels, and AtPGPs were detected with a polyclonal anti-PGP antibody on Western blots. (C) Coimmunoprecipitation of TWD1 using anti-AtPGP1 antiserum. Membranes from Arabidopsis cell suspension culture were cross-linked with DTBP, solubilized using 2% TX-100, and immunoprecipitated using anti-AtPGP1 antiserum. Immunoprecipitated proteins were separated by 12.5% (lane 1–3) and 7.5% PAGE (lane 4) and probed with anti-TWD1 (lane 1–3) and anti-AtPGP1 (lane 4) antiserum, respectively. As negative control, unspecific binding of proteins to empty protein G was monitored (lane 2). Note that the size difference of the coprecipitated wild-type TWD1 in lane 3 having a slightly smaller weight than the HA-epitope–tagged TWD1 run in parallel (lane 1) is due to the lack of the HA-epitope. Molecular size markers on the left and right correspond to lanes 1–3, respectively; positions of TWD1 and AtPGP1 are indicated.
To demonstrate the TWD1/AtPGP1 complex in vivo, AtPGP1 was immunoprecipitated from solubilized wild-type membrane microsomes after cross-linking with thiol-cleavable DTBP (Figure 6C). Cross-linking was used, because strong detergent treatment is essential to solubilize both proteins from the plasma membrane. These treatments were expected to disrupt protein-protein interaction (Weixel and Bradbury, 2000). To prevent contamination of the eluate with the heavy chain of rabbit anti-AtPGP1—running at similar size than TWD1—the primary antibody was additionally immobilized to protein G by cross-linking. The absence of heavy chain antibodies was verified using only secondary antibodies detecting no band even after prolonged exposure (unpublished data).
Wild-type TWD1, which is slightly smaller than HATWD1 (Figure 6C, lane 1), was indeed detectable (Figure 6C, lane 3) using anti-TWD1 antiserum, suggesting coprecipitation with AtPGP1 (Figure 6C, lane 4). This was the case under reducing conditions (+DTT), which cleave the DTBP cross-linker. The intensity of a TWD1 band under reducing conditions was approximately 10 times higher than under nonreducing conditions (unpublished data). This faint signal is not surprising as acidic elution of the complex can result in partial cleavage of sulfur double bonds of the cross-linker. No TWD1 could be detected in control experiments using empty protein G (Figure 6C, lane 2).
Polar Auxin Transport Is Reduced in twd1 and atpgp Mutants
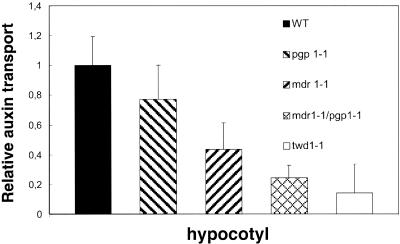
Measurements of polar auxin transport in hypocotyls of atpgp19-1 (atmdr1-1) and atpgp1-1 mutants shows reduction of auxin transport between 44 and 77%. This reduction in transport activity is more drastic in the double mutant atpgp1-1/atpgp19-1 (atmdr1-1), where the transport is reduced to 24% of wild-type plants. Polar auxin transport in twd1 mutants is reduced to 14% of wild-type plants even when no mutations in AtPGP1 and AtPGP19 are present (Figure 7). The methods used to measure auxin transport in this article represent a refinement of those used in Noh et al. (2001) (see MATERIALS AND METHODS).
Figure 7.
Relative auxin transport activity in hypocotyls of young seedlings The ABC transporter mutation atpgp1-1 exhibits slightly reduced polar auxin transport under these conditions shown, whereas the atpgp19-1 (atmdr1-1) exhibits markedly reduced polar auxin transport. Auxin transport in the double atpgp1-1/atpgp19-1 (atmdr1-1) mutant was dramatically impaired. The twd1-1 mutant showed even less polar auxin transport activity.
DISCUSSION
Mutations in the Arabidopsis TWD1 gene encoding a protein highly homologous to FK506-binding immunophilins cause a dramatic pleiotropic phenotype including epinastic cotyledons, a reduction of cell elongation, and a disoriented growth behavior (see Figure 1; B. Schulz, B. Saal, D. Wanke, M. Lafos, H.Ü. Kolukisaoglu, B.P. Dilkes, and K.A.J. Feldman, unpublished results). Because TWD1 does not exhibit PPIase activity (Kamphausen et al., 2002) or complement yeast FKBP12, we assumed that TWD1 action is mediated indirectly by protein-protein interaction. In a yeast two-hybrid screen we identified the Arabidopsis ABC transporter AtPGP1 as a TWD1-interacting partner. We provide several lines of evidence that AtPGP1 and AtPGP19 (AtMDR1) are relevant interacting partners of TWD1. Moreover, we collected indirect and direct proof that a functional complex of TWD1-AtPGP1/AtPGP19 is required for proper plant development: phenotypes observed for twd1 and the atpgp1-1/atpgp19-1 (mdr1-1) double mutant— but not of individual atpgp1-1 or atpgp19-1 mutants—resemble each other in early stages of development, and auxin transport is severely reduced in twd1-1 and atpgp1/atpgp19 mutant hypocotyls.
Interaction with AtPGP1 Is Mediated by the PPIase-like Domain of TWD1
Two-hybrid and in vitro analyses suggest that TWD1 specifically interacts with both AtPGP1 and AtPGP19 (AtMDR1). These ABC transporters were isolated by NPA-chromatography in a high-affinity NPA-binding fraction together with TWD1 (misannotated as a cyclophilin 5-like protein) and AtPGP2 as minority components (Noh et al., 2001; Murphy et al., 2002). TWD1 does not interact with the C-terminus of any other AtPGP in a common subbranch of a phylogenetic tree (Martinoia et al., 2002), which emphasizes the specificity of interaction of TWD1 with AtPGP1 and AtPGP19.
Mapping of TWD1 motifs required for AtPGP1 binding demonstrated that AtPGP1 recognizes the N-terminal PPI-ase-like domain. This is an unexpected finding as the TPR domain is usually implicated in protein-protein interactions as observed for other FKBPs (Lamb et al., 1995; Das et al., 1998; Harrar et al., 2001). However, another exception is known: hFKBP52 was found to interact with dynein, FAP48, and PHAX via its PPIase-like domain (Chambraud et al., 1996, 1999). Mapping analysis revealed that AtPGP19 in contradiction to AtPGP1 requires additional TWD1 regions for interaction. Thus far all attempts to measure any PPIase activity for TWD1 using synthetic peptide substrates have failed (Kamphausen et al., 2002). Additionally, TWD1 also failed to complement the yeast FKBP12 mutant (our unpublished results). Finally, we found no interference of FK506 in TWD1-AtPGP1 interactions by using either the yeast two-hybrid system or in vitro using TWD1-affinity chromatography.
These findings might not be surprising because 12 aa residues involved in high-affinity interactions between human FKBP12 and FK506 or rapamycin, which are highly conserved in most mammalian and yeast FKBPs (Van Duyne et al., 1991), are poorly conserved in most plant FKBPs. Only 4 of the 14 residues are conserved in TWD1 (74Y, 85F, 87D, 132Y). Interestingly, FKBP12 from Vicia faba (75% conservation) also failed to mediate FK506 and rapamycin action in yeast (Xu et al., 1998). Amino acid substitution presumably also abolishes drug binding and PPIase activity in human FKBP38 (Pedersen et al., 1999).
In summary, our results support the possibility that a modified PPIase-like domain, which is able to interact specifically with xenobiotic immunsuppressants in animal cells, could have acquired the ability to interact with ABC transporters, i.e., AtPGP1 and AtPGP19 in plants.
TWD1 Is a Unique Membrane-anchored FKBP Forming a Complex with AtPGP1 in the Plasma Membrane
Various members of the FKBP subfamily in plants are expressed as soluble proteins in different intracellular compartments including the cytoplasm, ER, and nucleus (Luan et al., 1996; Reddy et al., 1998). High-molecular-weight FKBP73 and FKBP77 from wheat have been identified as part of a HSP90 hetero-complex in vitro (Reddy et al., 1998).
A database search identified C-terminal putative transmembrane domains additionally only in human and mouse FKBP38 as well as in Drosophila FKBP45, but TWD1 is the first immunophilin shown to be a membrane-anchored protein. Anchoring of TWD1 on the plasma membrane was demonstrated by fractionation of membrane preparations. Only high concentrations of Triton X-100, but not gentle treatments, destroying hydrophilic or hydrophobic protein-protein or membrane-protein interactions, resulted in partial solubilization of the TWD1 protein (Figure 4A). The fact that solubilization was always incomplete indicates that additional interactions (e.g., to AtPGP1/19) might contribute to TWD1 retention on the plasma membrane.
Membrane fractionation and confocal laser microscopy of transgenic plants expressing a HA-tagged version of TWD1 demonstrate that TWD1 is indeed localized on the plasma membrane (Figures 4B and 5). Despite its low abundance, we were able to confirm these results by detection of TWD1 in purified plasma membranes from wild-type plant material derived from aqueous two-phase partitioning (Figure 4B). Weak background fluorescence surrounded the chloroplasts in the protoplasts examined (green stain in Figure 5C). Immunoblotting of thylakoids and chloroplast inner envelope membranes isolated from plant material overexpressing HA-TWD1 never showed any signals (our unpublished results), which excludes a chloroplast localization of TWD1. In contradiction to our data, Kamphausen et al. (2002) immunolocalized TWD1 to the plasma membrane as well as to the tonoplast in plants overexpressing HA-TWD1 using electron microscopy. In these plants but not in wild-type preparations, white unstructured areas were distinguishable that were discussed to occur from massive overproduction of a membrane protein.
On cross-linking, TWD1 could be detected in an immunoprecipitation assay using anti-AtPGP1. This result suggests a functional AtPGP1 and TWD1 protein complex on the plasma membrane (Figure 6C).
Possible Physiological Role of TWD1
Two plausible models offer explanations for the observed TWD1-AtPGP1/AtPGP19 interaction. The first hypothesis suggests that TWD1 is involved in proper folding or protein trafficking of AtPGP1 to the plasma membrane. Based on our results, this option can be excluded because membrane fractionation showed a very similar distribution of AtPGP1 in twd1-1 mutant and wild-type plants (our unpublished results).
The second model hypothesizes that immunophilin-like proteins play a regulatory role in multiprotein complexes. TWD1 could thus function as a potential regulator of AtPGP1 and AtPGP19 transport activities by means of domain-specific protein-protein interactions. This model is supported by several studies: FKBP12 is a subunit and inhibits basal signaling of two intracellular calcium release channels, the inositol 1,4,5-trisphosphate and ryanodin receptor (Cameron et al., 1995; Timerman et al., 1995). It has also been suggested that FKBP12 regulates MDR-like ABC transporters (Hemenway and Heitman, 1996; Mealey et al., 1999). Interestingly, FKBP12-dependent regulation of the ryanodine-sensitive Ca2+ channel and MDR3-mediated drug resistance is independent of FKBP12 PPIase activity (Timerman et al., 1995; Hemenway and Heitman, 1996). As TWD1 interacting clones of AtPGP1 and AtPGP19 cover the Walker A and B boxes of the C-terminal nucleotide binding fold involved in nucleotide binding and hydrolysis (Rea et al., 1998; Martinoia et al., 2002), it is conceivable that interaction of this domain with TWD1 affects ATP binding or hydrolysis.
The putative regulatory impact of TWD1 on AtPGP1 and AtPGP19 (AtMDR1) activity is hard to test at present because the in vivo substrate specificity of AtPGP1 and AtPGP19 (AtMDR1) must still be resolved (Thomas et al., 2000; Noh et al., 2001; Windsor et al., 2003). However, AtPGP1 and AtPGP19 have a direct or indirect role in polar auxin transport (Luschnig 2002) and the structural similarity between auxin and synthetic indolic substrates transported by mammalian MDR proteins is obvious (Nelson et al., 1998). Measurements of polar auxin transport in hypocotyls of the ABC transporter mutant atpgp1-1 show a slight and somewhat variable reduction in transport, whereas auxin transport in atmdr1-1 (atpgp19) is clearly reduced. The reduction of auxin transport is even more pronounced in the atpgp1-1/atpgp19-1 (atmdr1-1) double mutant and could be interpreted as an additive effect of both mutations. The mutation in TWD1 results in even greater reduction of polar auxin transport in hypocotyls, which indicates that TWD1 is necessary for the activity and interaction of both ABC transporters. A role for TWD1 in auxin transport regulation is also suggested by unsuccessful attempts to measure auxin transport in an AtPGP19 (AtMDR1) expressing heterologous yeast system (Noh et al., 2001; Murphy, unpublished results). TWD1 may be a specific regulator of putative auxin transporters or possibly also MDR-like ABC transporters (Luschnig, 2002; Muday and Murphy, 2002). Such an interaction is further suggested by recent binding studies of Arabidopsis plasma membrane proteins to a matrix containing an immobilized form of the polar auxin transport inhibitor NPA (Murphy et al., 2002). In these studies, AtPGP1, AtPGP19 (AtMDR1), and small amounts of TWD1 copurified in the same NPA-binding fraction (fraction IV). We could corroborate these data by modifying the experimental setup. By binding HA-epitope–tagged TWD1 protein that was isolated from plant material in the presence of NPA to an anti-HA-epitope matrix, we were able to show that AtPGPs solubilized from microsomal protein preparations bound to the TWD1 protein. This interaction could be disrupted by treatments with NPA. Isolating TWD1 from microsomal preparations was successful only in the absence of NPA (Figure 6).
Importantly, in the previous NPA-affinity matrix experiments, only residual amounts of TWD1 were retained when the NPA used was immobilized via a critical carboxylic acid essential to its function as an auxin transport inhibitor (Murphy et al., 2002). In the experiments described herein, elution with conjugated, and therefore physiologically active, NPA appeared to disrupt all TWD1-AtPGP interactions. This again, shows a specific and NPA-sensitive interaction of TWD1 with AtPGPs in the plant cell.
It is tempting to speculate that the similar phenotypes of twd1-1 and atpgp1-1/atpgp19-1 double mutants (Figure 1) are both the result of abnormal distribution of growth factors such as auxin. Strong growth effects on leaf lamina expansion (epinastic growth) and root length reduction support this assumption. If so, the absence of TWD1 would not allow the formation of a transport-competent complex, which involves ABC transporters of the AtPGP/AtMDR-subfamily.
Acknowledgments
We thank Drs. H. Sze, A. Vitale, and R. Serrano for providing antisera; J. Kudla for CBL1 and CIPK; K. Palme for Y97; and M.G. Palmgren for pMP900 plasmids. We are grateful to M. Meylan-Bettex and Dr. L. Bovet for the preparation of Arabidopsis thylakoid and inner envelope membranes and V. Vincenzetti for excellent technical assistance. We thank Drs. U.I. Flügge for continuous support, E. Spalding for sharing results before publication, and Drs. N. Johnsson and R. Schmidt for critical reading of the manuscript. This work was supported by the Swiss National Foundation, Deutsche Forschungsgemeinschaft (Schu/821–2), the European Community (LATIN, BIOTEC 4), and the Ministerium für Schule, Wissenschaft, und Forschung des Landes NRW, Novartis, and the Alexander von Humboldt-Foundation (Feodor-Lynen and Novartis fellowships to M.G.). A.S.M. and J.J.B. were supported by the National Science Foundation (NSF Grant 0132803).
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.E02–10–0698. Article and publication date are available at www.molbiolcell.org/cgi/doi/10.1091/mbc.E02-10-0698.
Abbreviations used: FKBPs, FK506-binding proteins; PPIases, cis-trans-peptidylprolyl isomerases; MDR, multidrug resistance; CsA, cyclosporin A; TPR, tetratrico-peptide repeat; twd1, twisted dwarf1; ABC, ATP-binding cassette; β-gal, β-galactosidase; PGP, P-glycoprotein; NPA, naphthylphthalamic acid.
References
- Aghdasi, B., Ye, K., Resnick, A., Huang, A., Ha, H.C., Guo, X., Dawson, T.M., Dawson, V.L., and Snyder, S.H. (2001). FKBP12, the 12-kDa FK506-binding protein, is a physiological regulator of the cell cycle. Proc. Natl. Acad. Sci. USA. 98, 2425–2430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arabidopsis Genome Initiative. (2000). Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408, 796–815. [DOI] [PubMed] [Google Scholar]
- Axelos, M., Curie, C., Mazzolini, L., Bardet, C., and Lescure, B. (1992). A protocol for transient gene expression in Arabidopsis thaliana protoplasts isolated from cell suspension cultures. Plant Physiol. Biochem. 30, 123–128. [Google Scholar]
- Berardini, T.Z., Bollmann, K., Sun, H., and Poethig, S. (2001). Regulation of vegetative phase change in Arabidopsis thaliana by cyclophilin 40. Science 291, 2405–2407. [DOI] [PubMed] [Google Scholar]
- Cameron, A.M., Steiner, J.P., Roskams, A.J., Ali, S.M., Ronnett, G.V., and Snyder, S.H. (1995). Calcineurin associated with the inositol 1,4,5-trisphosphate receptor-FKBP12 complex modulates Ca2+ flux. Cell 83, 463–472. [DOI] [PubMed] [Google Scholar]
- Cardenas, M.E., Hemenway, C., Muir, R.S., Ye, R., Fiorentino, D., and Heitman, J. (1994). Immunophilins interact with calcineurin in the absence of exogenous immunosuppressive ligands. EMBO J. 13, 5944–5957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardenas, M.E., Cruz, M.C., Del Poeta, M., Chung, N., Perfect, J.R., and Heitman, J. (1999). Antifungal activities of antineoplastic agents: Saccharomyces cerevisiae as a model system to study drug action. Clin. Microbiol. Rev. 12, 583–611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambraud, B., Radanyi, C., Camonis, J.H., Shazand, K., Rajkowski, K., and Baulieu, E.E. (1996). FAP48, a new protein that forms specific complexes with both immunophilins FKBP59 and FKBP12. Prevention by the immunosuppressant drugs FK506 and rapamycin. J. Biol. Chem. 271, 32923–32929. [DOI] [PubMed] [Google Scholar]
- Chambraud, B., Radanyi, C., Camonis, J.H., Rajkowski, K., Schumacher, M., and Baulieu, E.E. (1999). Immunophilins, Refsum disease, and lupus nephritis: the peroxisomal enzyme phytanoyl-COA alpha-hydroxylase is a new FKBP-associated protein. Proc. Natl. Acad. Sci. USA 96, 2104–2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das, A.K., Cohen, P.T.W., and Barford, D. (1998). The structure of the tetratrico-peptide repeats of protein phosphatase 5, implications for the TPR-mediated protein-protein interactions. EMBO J. 17, 1192–1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolinski, K., Muir, S., Cardenas, M., and Heitman, J. (1997). All cyclophilins and FK506 binding proteins are, individually and collectively, dispensable for viability in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 94, 13093–13098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudler, R., and Hertig, C. (1992). Structure of an mdr-like gene from Arabidopsis thaliana J. Biol. Chem. 267, 5882–5888. [PubMed] [Google Scholar]
- Faure, J.D., Vittorioso, P., Santoni, V., Fraisier, V., Prinsen, E., Barlier, I., Van Onckelen, H., Caboche, M., and Bellini, C. (1998). The PASTICCINO genes of Arabidopsis thaliana are involved in the control of cell division and differentiation. Development 125, 909–918. [DOI] [PubMed] [Google Scholar]
- Fuglsang, A.T., Visconti, S., Drumm, K., Jahn, T., Stensballe, A., Mattei, B., Jensen, O.N., Aducci, P., and Palmgren, M.G. (1999). Binding of 14-3-3 protein to the plasma membrane H(+)-ATPase AHA2 involves the three C-terminal residues Tyr(946)-Thr-Val and requires phosphorylation of Thr(947). J. Biol. Chem. 274, 36774–36780. [DOI] [PubMed] [Google Scholar]
- Geisler, M., Frangne, N., Gomès, E., Martinoia, E., and Palmgren, M.G. (2000). The ACA4 gene of Arabidopsis encodes a vacuolar membrane calcium pump that improves salt tolerance in yeast. Plant Phys. 124, 1814–1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrar, Y., Bellini, C., and Faure, J.D. (2001). FKBPs: at the crossroads of folding and transduction. Trends Plant Sci. 6, 426–431. [DOI] [PubMed] [Google Scholar]
- Hemenway, C.S., and Heitman, J. (1996). Immunosuppressant target protein FKBP12 is required for P-glycoprotein function in yeast. J. Biol. Chem. 271, 18527–18534. [DOI] [PubMed] [Google Scholar]
- Kamphausen, T., Fanghänel, J., Neumann, D., Schulz, B., and Rahfeld, J.-U. (2002). Characterisation of Arabidopsis thaliana AtFKBP42 that is membrane bound and interacts with Hsp90. Plant J. 32, 263–276. [DOI] [PubMed] [Google Scholar]
- Lamb, J.R., Tugendreich, S., and Hieter, P. (1995). Tetratrico-peptide repeat interactions: to TPR or not to TPR? Trends Biochem. Sci. 20, 257–259. [DOI] [PubMed] [Google Scholar]
- Luan, S., Kudla, J., Gruissem, W., and Schreiber, S.L. (1996). Molecular characterization of a FKBP-type immunophilin from higher plants. Proc. Natl. Acad. Sci. USA 93, 6964–6969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luschnig, C. (2002). Auxin transporters: ABC transporters join the club. Trends Plant Sci. 7, 329–333. [DOI] [PubMed] [Google Scholar]
- Martinoia, E., Klein, M., Geisler, M., Bovet, L., Forestier, C., Kolukisaoglu, Ü., Mueller-Roeber, B., and Schulz, B. (2002). Multifunctionality of plant ABC transporters—more than just detoxifiers. Planta 214, 345–355. [DOI] [PubMed] [Google Scholar]
- Mealey, K.L., Barhoumi, R., Burghardt, R.C., McIntyre, B.S., Sylvester, P.W., Hosick, H.L., and Kochevar, D.T. (1999). Immunosuppressant inhibition of P-glycoprotein function is independent of drug-induced suppression of peptide-prolyl isomerase and calcineurin activity. Cancer Chemother. Pharmacol. 44, 152–158. [DOI] [PubMed] [Google Scholar]
- Muday, G.K., and Murphy, A.S. (2002). An emerging model of auxin transport regulation. Plant Cell 14, 293–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy, A.S., Peer, W.A., and Taiz, L. (2000). Regulation of auxin transport by aminopeptidases and endogenous flavonoids. Planta 211, 315–324. [DOI] [PubMed] [Google Scholar]
- Murphy, A.S., Hoogner, K.R., Peer, W.A., and Taiz, L. (2002). Identification, purification and molecular cloning of N-1-naphthylphthalmic acid-binding plasma membrane-associated aminopeptidases from Arabidopsis. Plant Phys. 128, 935–950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson, E.J., Zinkin, N.T., and Hinkle, P.M. (1998). Fluorescence methods to assess multidrug resistance in individual cells. Cancer Chemother. Pharmacol. 42, 292–299. [DOI] [PubMed] [Google Scholar]
- Németh, K. et al. (1998). Pleiotropic control of glucose and hormone responses by PRL1, a nuclear WD protein, in Arabidopsis. Genes Dev. 12, 3059–3073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noh, B., Murphy, A.S., and Spalding, E.P. (2001). Multidrug resistance-like genes of. Arabidopsis required for auxin transport and auxin-mediated development. Plant Cell 13, 2441–2454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owens-Grillo, J.K., Stancato, L.F., Hoffmann, K., Pratt, W.B., and Krishna, P. (1996). Binding of immunophilins to the 90 kDa heat shock protein (hsp90) via a tetratrico-peptide repeat domain is a conserved protein interaction on plants. Biochemistry 35, 15249–15255. [DOI] [PubMed] [Google Scholar]
- Pedersen, K.M., Finsen, B. Celis, J.E., and Jensen, N.A. (1999). muFKBP 38, A novel murine immunophilin homolog differentially expressed in Schwannoma cells and central nervous system neurons in vivo. Electrophoresis 20, 249–255. [DOI] [PubMed] [Google Scholar]
- Pratt, W.B., Krishna, P., and Olsen, L.J. (2001). Hsp90-binding immunophilins in plants: the protein movers. Trends Plant Sci. 6, 54–58. [DOI] [PubMed] [Google Scholar]
- Rea, A.P., Ze-Sheng, L., Yu-Ping, L., Drozdowicz, Y.M., and Martinoia, E. (1998). From vacuolar GS-X pumps to multispecific ABC transporters. Annu. Rev. Plant Physiol. 49, 727–760. [DOI] [PubMed] [Google Scholar]
- Reddy, R.K., Kurek, I., Silverstein, A.M., Chinkers, M., Breiman, A., and Krishna, P. (1998). High-molecular-weight FK506-binding proteins are components of heat-shock protein 90 heterocomplexes in wheat germ lysate. Plant Physiol. 118, 1395–1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Fernandez, R., Davies, T.G.E., Coleman, J.O.D., and Rea, P.A. (2001). The Arabidopsis thaliana ABC protein superfamily, a complete inventory. J. Biol. Chem. 276, 30231–30244. [DOI] [PubMed] [Google Scholar]
- Schiene, C., and Fischer, G. (2000). Enzymes that catalyse the restructuring of proteins. Curr. Opin. Struct. Biol. 10, 40–45. [DOI] [PubMed] [Google Scholar]
- Schubert, M., Petersson, U.A., Haas, B.J., Funk, C., Schroder, W.P., and Kieselbach, T. (2002). Proteome map of the chloroplast lumen of Arabidopsis thaliana. J. Biol. Chem. 277, 8354–65. [DOI] [PubMed] [Google Scholar]
- Shi, J., Kim, K.N., Ritz, O., Albrecht, V., Gupta, R., Harter, K., Luan, S., and Kudla, J. (1999). Novel protein kinases associated with calcineurin B-like calcium sensors in Arabidopsis. Plant Cell 11, 2393–2406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidler, M., Hassa, P., Hasan, S., Ringli, C., and Dudler, R. (1998). Involvement of an ABC transporter in a developmental pathway regulating hypocotyl cell elongation in the light. Plant Cell 10, 1623–1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverstein, A.M., Galigniana, M.D., Kanelakis, K.C., Radanyi, C., Renoir, J.M., and Pratt, W.B. (1999). Different regions of the immunophilin FKBP52 determine its association with the glucocorticoid receptor, hsp90, and cytoplasmic dynein. J. Biol. Chem. 274, 36980–36986. [DOI] [PubMed] [Google Scholar]
- Thomas, C., Rajagopal, A., Windsor, B., Dudler, R., Lloyd, A., and Roux, S.J. (2000). A role for ectophosphatase in xenobiotic resistance. Plant Cell 12, 519–533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timerman, A.P., Wiederrecht, G., Marcy, A., and Fleischer, S. (1995). Characterization of an exchange reaction between soluble FKBP-12 and the FKBP ryanodine receptor complex. Modulation by FKBP mutants deficient in peptidyl-prolyl isomerase activity. J. Biol. Chem. 270, 2451–2459. [DOI] [PubMed] [Google Scholar]
- Überlacker, B., and Werr, W. (1996). Vectors with rare-cutter restriction enzyme sites for expression of open reading frames in transgenic plants. Mol. Breeding 2, 293–295. [Google Scholar]
- Vittorioso, P., Cowling, R., Faure, J.D., Caboche, M., and Bellini, C. (1998). Mutation in the Arabidopsis PASTICCINO1 gene, which encodes a new FK506-binding protein-like protein, has a dramatic effect on plant development. Mol. Cell Biol. 18, 3034–3043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Duyne, G.D., Staendaert, R.F., Karplus, P.A., Schreiber, S.L., and Clardy, J. (1991). Atomic structure of FKBP-FK506, an immunophilin-ligand complex. Science 252, 839–842. [DOI] [PubMed] [Google Scholar]
- Weixel, K.M., and Bradbury, N.A. (2000). The carboxyl terminus of the cystic fibrosis transmembrane conductance regulator binds to AP-2 clathrin adaptors. J. Biol. Chem. 275, 3655–3660. [DOI] [PubMed] [Google Scholar]
- Windsor, B., Roux, S.J., and Lloyd, A. (2003). Multiherbicide tolerance conferred by AtPGP1 and apyrase overexpression in Arabidopsis thaliana. Nat. Biotech. 21, 428–433. [DOI] [PubMed] [Google Scholar]
- Xu, Q., Liang, S., Kudla, J., and Luan, S. (1998). Molecular characterization of a plant FKBP12 that does not mediate action of FK506 and rapamycin. Plant J. 15, 511–519. [DOI] [PubMed] [Google Scholar]