Abstract
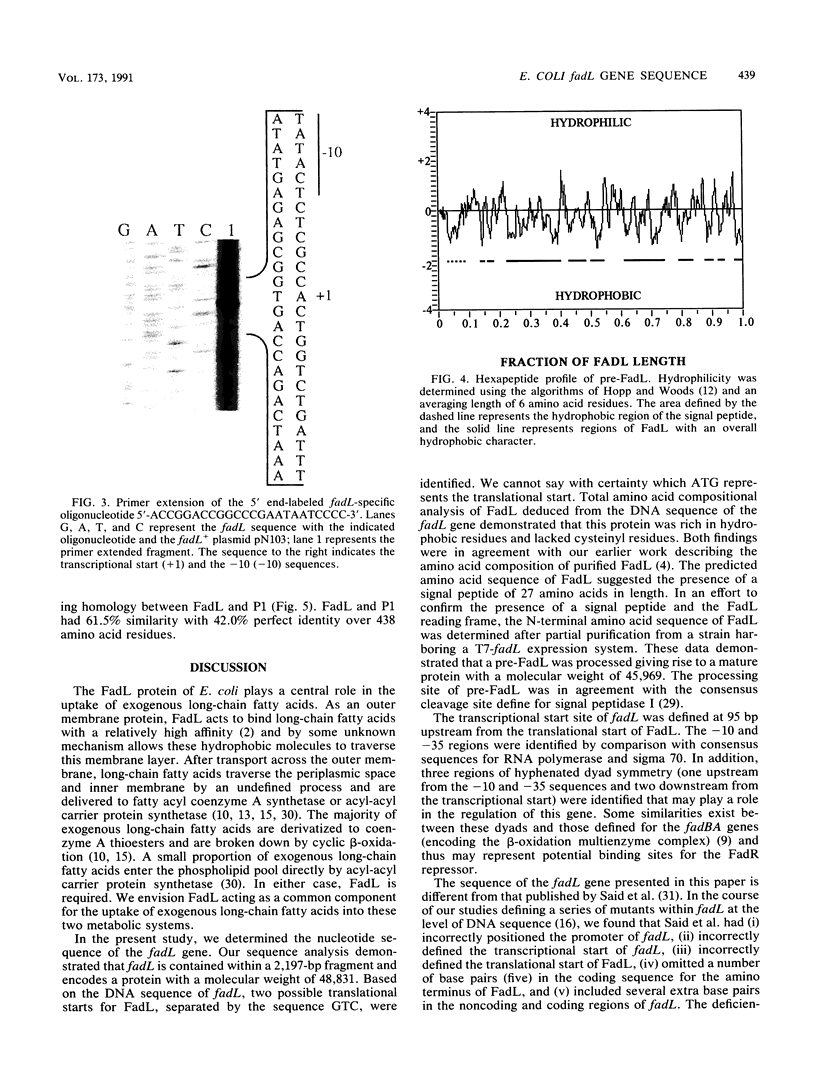
The fadL gene of Escherichia coli encodes an outer membrane protein (FadL) that plays a central role in the uptake of exogenous long-chain fatty acids. The nucleotide sequence of the fadL gene revealed a single open reading frame of 1,344 bp encoding a protein with 448 amino acid residues and a molecular weight of 48,831. The transcriptional start, analyzed by primer extension, was shown to be 95 bp upstream from the translational start. Apparent -10 and -35 regions were found at -12 and -37 bp upstream from the transcriptional start. Three regions with hyphenated dyad symmetry (two between the transcriptional start and the translational start and one upstream from the -10 and -35 regions) were identified that may play a role in the expression of fadL. The protein product of the fadL gene contained a signal sequence and signal peptidase I cleavage site similar to that defined for other E. coli outer membrane proteins. The N-terminal sequence of mature FadL protein was determined by automated amino acid sequencing of protein purified from the outer membrane of a strain harboring fadL under the control of a T7 RNA polymerase-responsive promoter. This amino acid sequence, Ala-Gly-Phe-Gln-Leu-Asn-Glu-Phe-Ser-Ser, verified the signal peptidase I cleavage site on pre-FadL and confirmed the N-terminal amino acid sequence of FadL predicted from the DNA sequence. Mature FadL contained 421 amino acid residues, giving a molecular weight of 45,969. The amino acid composition of FadL deduced from the DNA sequence suggested that this protein contained an abundance of hydrophobic amino acid residues and lacked cysteinyl residues. The hydrophobic amino acids within FadL were predicted to contribute to at least five regions of the protein with an overall hydrophobic character. The amino acid sequence of FadL was used to search GenBank for other proteins with amino acid sequence homology. These data demonstrated that FadL and the heat-modifiable outer membrane protein P1 of Haemophilus influenzae type b were 60.5% conserved and 42.0% identical over 438 amino acid residues.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Black P. N. Characterization of FadL-specific fatty acid binding in Escherichia coli. Biochim Biophys Acta. 1990 Aug 28;1046(1):97–105. doi: 10.1016/0005-2760(90)90099-j. [DOI] [PubMed] [Google Scholar]
- Black P. N., Kianian S. F., DiRusso C. C., Nunn W. D. Long-chain fatty acid transport in Escherichia coli. Cloning, mapping, and expression of the fadL gene. J Biol Chem. 1985 Feb 10;260(3):1780–1789. [PubMed] [Google Scholar]
- Black P. N., Said B., Ghosn C. R., Beach J. V., Nunn W. D. Purification and characterization of an outer membrane-bound protein involved in long-chain fatty acid transport in Escherichia coli. J Biol Chem. 1987 Jan 25;262(3):1412–1419. [PubMed] [Google Scholar]
- Black P. N. The fadL gene product of Escherichia coli is an outer membrane protein required for uptake of long-chain fatty acids and involved in sensitivity to bacteriophage T2. J Bacteriol. 1988 Jun;170(6):2850–2854. doi: 10.1128/jb.170.6.2850-2854.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dagert M., Ehrlich S. D. Prolonged incubation in calcium chloride improves the competence of Escherichia coli cells. Gene. 1979 May;6(1):23–28. doi: 10.1016/0378-1119(79)90082-9. [DOI] [PubMed] [Google Scholar]
- Devereux J., Haeberli P., Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 1):387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiRusso C. C. Primary sequence of the Escherichia coli fadBA operon, encoding the fatty acid-oxidizing multienzyme complex, indicates a high degree of homology to eucaryotic enzymes. J Bacteriol. 1990 Nov;172(11):6459–6468. doi: 10.1128/jb.172.11.6459-6468.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frerman F. E., Bennett W. Studies on the uptake of fatty acids by Escherichia coli. Arch Biochem Biophys. 1973 Nov;159(1):434–443. doi: 10.1016/0003-9861(73)90471-2. [DOI] [PubMed] [Google Scholar]
- Hanson M. S., Cope L. D., Hansen E. J. Expression of the heat-modifiable major outer membrane protein of Haemophilus influenzae type b is unrelated to virulence. Infect Immun. 1989 Jun;57(6):1639–1646. doi: 10.1128/iai.57.6.1639-1646.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopp T. P., Woods K. R. Prediction of protein antigenic determinants from amino acid sequences. Proc Natl Acad Sci U S A. 1981 Jun;78(6):3824–3828. doi: 10.1073/pnas.78.6.3824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kameda K., Nunn W. D. Purification and characterization of acyl coenzyme A synthetase from Escherichia coli. J Biol Chem. 1981 Jun 10;256(11):5702–5707. [PubMed] [Google Scholar]
- Kassavetis G. A., Geiduschek E. P. Bacteriophage T4 late promoters: mapping 5' ends of T4 gene 23 mRNAs. EMBO J. 1982;1(1):107–114. doi: 10.1002/j.1460-2075.1982.tb01132.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein K., Steinberg R., Fiethen B., Overath P. Fatty acid degradation in Escherichia coli. An inducible system for the uptake of fatty acids and further characterization of old mutants. Eur J Biochem. 1971 Apr;19(3):442–450. doi: 10.1111/j.1432-1033.1971.tb01334.x. [DOI] [PubMed] [Google Scholar]
- Loeb M. R. Protection of infant rats from Haemophilus influenzae type b infection by antiserum to purified outer membrane protein a. Infect Immun. 1987 Nov;55(11):2612–2618. doi: 10.1128/iai.55.11.2612-2618.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maloy S. R., Ginsburgh C. L., Simons R. W., Nunn W. D. Transport of long and medium chain fatty acids by Escherichia coli K12. J Biol Chem. 1981 Apr 25;256(8):3735–3742. [PubMed] [Google Scholar]
- Matsudaira P. Sequence from picomole quantities of proteins electroblotted onto polyvinylidene difluoride membranes. J Biol Chem. 1987 Jul 25;262(21):10035–10038. [PubMed] [Google Scholar]
- Messing J. New M13 vectors for cloning. Methods Enzymol. 1983;101:20–78. doi: 10.1016/0076-6879(83)01005-8. [DOI] [PubMed] [Google Scholar]
- Munson R., Jr, Grass S. Purification, cloning, and sequence of outer membrane protein P1 of Haemophilus influenzae type b. Infect Immun. 1988 Sep;56(9):2235–2242. doi: 10.1128/iai.56.9.2235-2242.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura K., Mizushima S. Effects of heating in dodecyl sulfate solution on the conformation and electrophoretic mobility of isolated major outer membrane proteins from Escherichia coli K-12. J Biochem. 1976 Dec;80(6):1411–1422. doi: 10.1093/oxfordjournals.jbchem.a131414. [DOI] [PubMed] [Google Scholar]
- Nunn W. D. A molecular view of fatty acid catabolism in Escherichia coli. Microbiol Rev. 1986 Jun;50(2):179–192. doi: 10.1128/mr.50.2.179-192.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunn W. D., Simons R. W., Egan P. A., Maloy S. R. Kinetics of the utilization of medium and long chain fatty acids by mutant of Escherichia coli defective in the fadL gene. J Biol Chem. 1979 Sep 25;254(18):9130–9134. [PubMed] [Google Scholar]
- Nunn W. D., Simons R. W. Transport of long-chain fatty acids by Escherichia coli: mapping and characterization of mutants in the fadL gene. Proc Natl Acad Sci U S A. 1978 Jul;75(7):3377–3381. doi: 10.1073/pnas.75.7.3377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliver D. Protein secretion in Escherichia coli. Annu Rev Microbiol. 1985;39:615–648. doi: 10.1146/annurev.mi.39.100185.003151. [DOI] [PubMed] [Google Scholar]
- Rock C. O., Jackowski S. Pathways for the incorporation of exogenous fatty acids into phosphatidylethanolamine in Escherichia coli. J Biol Chem. 1985 Oct 15;260(23):12720–12724. [PubMed] [Google Scholar]
- Said B., Ghosn C. R., Vu L., Nunn W. D. Nucleotide sequencing and expression of the fadL gene involved in long-chain fatty acid transport in Escherichia coli. Mol Microbiol. 1988 May;2(3):363–370. doi: 10.1111/j.1365-2958.1988.tb00040.x. [DOI] [PubMed] [Google Scholar]
- Sallus L., Haselbeck R. J., Nunn W. D. Regulation of fatty acid transport in Escherichia coli: analysis by operon fusion. J Bacteriol. 1983 Sep;155(3):1450–1454. doi: 10.1128/jb.155.3.1450-1454.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shine J., Dalgarno L. Determinant of cistron specificity in bacterial ribosomes. Nature. 1975 Mar 6;254(5495):34–38. doi: 10.1038/254034a0. [DOI] [PubMed] [Google Scholar]
- Studier F. W., Moffatt B. A. Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J Mol Biol. 1986 May 5;189(1):113–130. doi: 10.1016/0022-2836(86)90385-2. [DOI] [PubMed] [Google Scholar]
- de Crombrugghe B., Busby S., Buc H. Cyclic AMP receptor protein: role in transcription activation. Science. 1984 May 25;224(4651):831–838. doi: 10.1126/science.6372090. [DOI] [PubMed] [Google Scholar]
- de Geus P., van Die I., Bergmans H., Tommassen J., de Haas G. Molecular cloning of pldA, the structural gene for outer membrane phospholipase of E. coli K12. Mol Gen Genet. 1983;190(1):150–155. doi: 10.1007/BF00330338. [DOI] [PubMed] [Google Scholar]



