Abstract
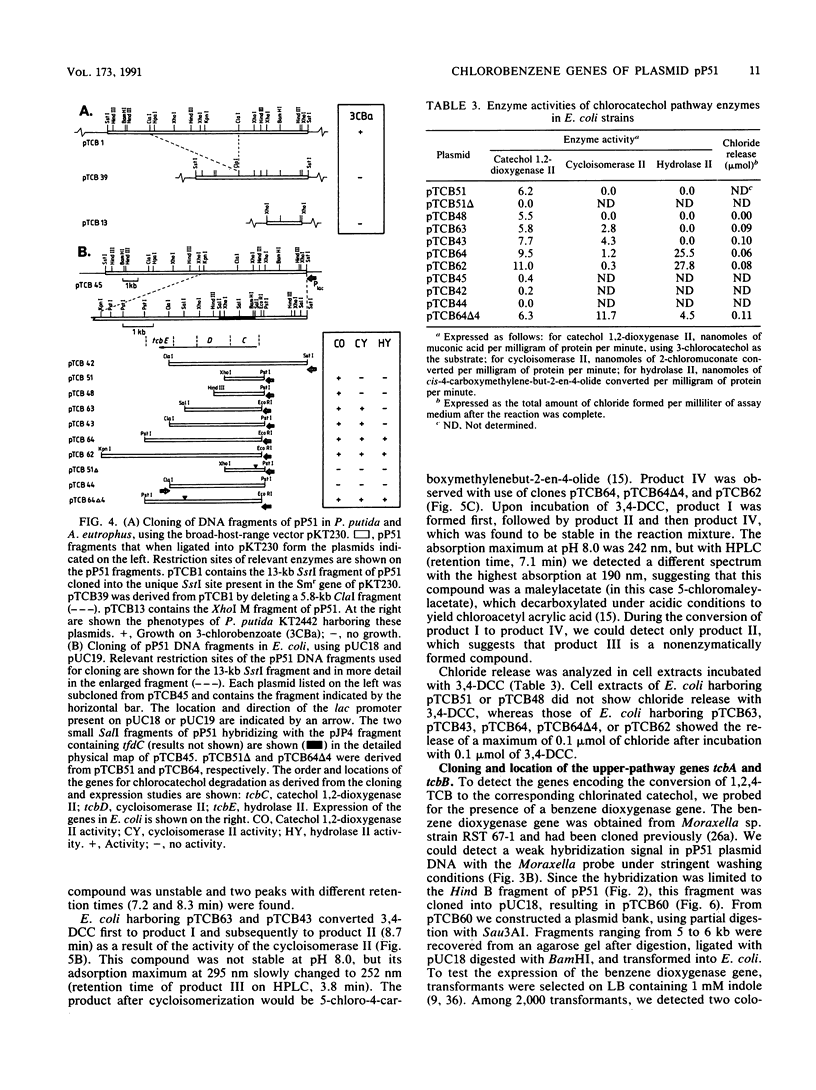
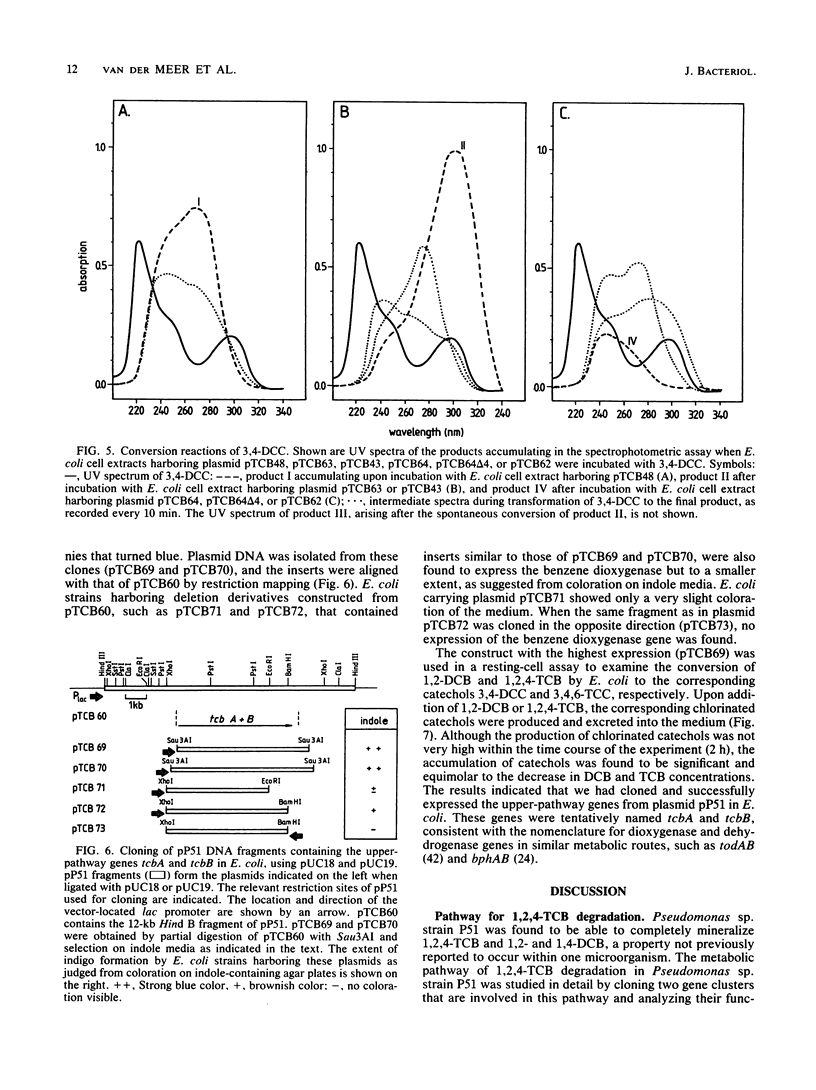
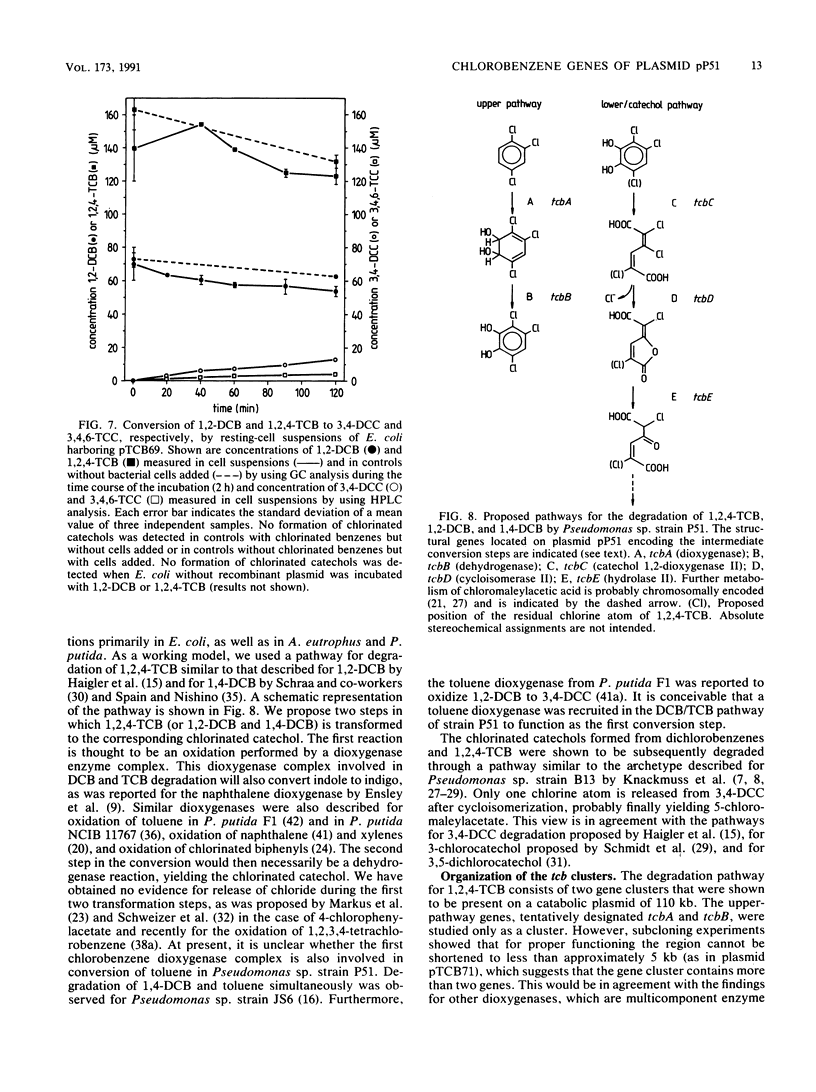
Pseudomonas sp. strain P51 is able to use 1,2-dichlorobenzene, 1,4-dichlorobenzene, and 1,2,4-trichlorobenzene as sole carbon and energy sources. Two gene clusters involved in the degradation of these compounds were identified on a catabolic plasmid, pP51, with a size of 110 kb by using hybridization. They were further characterized by cloning in Escherichia coli, Pseudomonas putida KT2442, and Alcaligenes eutrophus JMP222. Expression studies in these organisms showed that the upper-pathway genes (tcbA and tcbB) code for the conversion of 1,2-dichlorobenzene and 1,2,4-trichlorobenzene to 3,4-dichlorocatechol and 3,4,6-trichlorocatechol, respectively, by means of a dioxygenase system and a dehydrogenase. The lower-pathway genes have the order tcbC-tcbD-tcbE and encode a catechol 1,2-dioxygenase II, a cycloisomerase II, and a hydrolase II, respectively. The combined action of these enzymes degrades 3,4-dichlorocatechol and 3,4,6-trichlorocatechol to a chloromaleylacetic acid. The release of one chlorine atom from 3,4-dichlorocatechol takes place during lactonization of 2,3-dichloromuconic acid.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bagdasarian M., Lurz R., Rückert B., Franklin F. C., Bagdasarian M. M., Frey J., Timmis K. N. Specific-purpose plasmid cloning vectors. II. Broad host range, high copy number, RSF1010-derived vectors, and a host-vector system for gene cloning in Pseudomonas. Gene. 1981 Dec;16(1-3):237–247. doi: 10.1016/0378-1119(81)90080-9. [DOI] [PubMed] [Google Scholar]
- Bradford M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976 May 7;72:248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- Burlage R. S., Hooper S. W., Sayler G. S. The TOL (pWW0) catabolic plasmid. Appl Environ Microbiol. 1989 Jun;55(6):1323–1328. doi: 10.1128/aem.55.6.1323-1328.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterjee D. K., Chakrabarty A. M. Restriction mapping of a chlorobenzoate degradative plasmid and molecular cloning of the degradative genes. Gene. 1984 Feb;27(2):173–181. doi: 10.1016/0378-1119(84)90138-0. [DOI] [PubMed] [Google Scholar]
- Ditta G., Stanfield S., Corbin D., Helinski D. R. Broad host range DNA cloning system for gram-negative bacteria: construction of a gene bank of Rhizobium meliloti. Proc Natl Acad Sci U S A. 1980 Dec;77(12):7347–7351. doi: 10.1073/pnas.77.12.7347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Don R. H., Weightman A. J., Knackmuss H. J., Timmis K. N. Transposon mutagenesis and cloning analysis of the pathways for degradation of 2,4-dichlorophenoxyacetic acid and 3-chlorobenzoate in Alcaligenes eutrophus JMP134(pJP4). J Bacteriol. 1985 Jan;161(1):85–90. doi: 10.1128/jb.161.1.85-90.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorn E., Knackmuss H. J. Chemical structure and biodegradability of halogenated aromatic compounds. Substituent effects on 1,2-dioxygenation of catechol. Biochem J. 1978 Jul 15;174(1):85–94. doi: 10.1042/bj1740085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorn E., Knackmuss H. J. Chemical structure and biodegradability of halogenated aromatic compounds. Two catechol 1,2-dioxygenases from a 3-chlorobenzoate-grown pseudomonad. Biochem J. 1978 Jul 15;174(1):73–84. doi: 10.1042/bj1740073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ensley B. D., Ratzkin B. J., Osslund T. D., Simon M. J., Wackett L. P., Gibson D. T. Expression of naphthalene oxidation genes in Escherichia coli results in the biosynthesis of indigo. Science. 1983 Oct 14;222(4620):167–169. doi: 10.1126/science.6353574. [DOI] [PubMed] [Google Scholar]
- Franklin F. C., Bagdasarian M., Bagdasarian M. M., Timmis K. N. Molecular and functional analysis of the TOL plasmid pWWO from Pseudomonas putida and cloning of genes for the entire regulated aromatic ring meta cleavage pathway. Proc Natl Acad Sci U S A. 1981 Dec;78(12):7458–7462. doi: 10.1073/pnas.78.12.7458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frantz B., Chakrabarty A. M. Organization and nucleotide sequence determination of a gene cluster involved in 3-chlorocatechol degradation. Proc Natl Acad Sci U S A. 1987 Jul;84(13):4460–4464. doi: 10.1073/pnas.84.13.4460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furukawa K., Miyazaki T. Cloning of a gene cluster encoding biphenyl and chlorobiphenyl degradation in Pseudomonas pseudoalcaligenes. J Bacteriol. 1986 May;166(2):392–398. doi: 10.1128/jb.166.2.392-398.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosal D., You I. S. Nucleotide homology and organization of chlorocatechol oxidation genes of plasmids pJP4 and pAC27. Mol Gen Genet. 1988 Jan;211(1):113–120. doi: 10.1007/BF00338401. [DOI] [PubMed] [Google Scholar]
- Ghosal D., You I. S. Operon structure and nucleotide homology of the chlorocatechol oxidation genes of plasmids pJP4 and pAC27. Gene. 1989 Nov 30;83(2):225–232. doi: 10.1016/0378-1119(89)90108-x. [DOI] [PubMed] [Google Scholar]
- Haigler B. E., Nishino S. F., Spain J. C. Degradation of 1,2-dichlorobenzene by a Pseudomonas sp. Appl Environ Microbiol. 1988 Feb;54(2):294–301. doi: 10.1128/aem.54.2.294-301.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haigler B. E., Spain J. C. Degradation of p-chlorotoluene by a mutant of Pseudomonas sp. strain JS6. Appl Environ Microbiol. 1989 Feb;55(2):372–379. doi: 10.1128/aem.55.2.372-379.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanahan D. Studies on transformation of Escherichia coli with plasmids. J Mol Biol. 1983 Jun 5;166(4):557–580. doi: 10.1016/s0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- Hansen J. B., Olsen R. H. Isolation of large bacterial plasmids and characterization of the P2 incompatibility group plasmids pMG1 and pMG5. J Bacteriol. 1978 Jul;135(1):227–238. doi: 10.1128/jb.135.1.227-238.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harayama S., Rekik M., Timmis K. N. Genetic analysis of a relaxed substrate specificity aromatic ring dioxygenase, toluate 1,2-dioxygenase, encoded by TOL plasmid pWW0 of Pseudomonas putida. Mol Gen Genet. 1986 Feb;202(2):226–234. doi: 10.1007/BF00331641. [DOI] [PubMed] [Google Scholar]
- Kukor J. J., Olsen R. H., Siak J. S. Recruitment of a chromosomally encoded maleylacetate reductase for degradation of 2,4-dichlorophenoxyacetic acid by plasmid pJP4. J Bacteriol. 1989 Jun;171(6):3385–3390. doi: 10.1128/jb.171.6.3385-3390.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markus A., Krekel D., Lingens F. Purification and some properties of component A of the 4-chlorophenylacetate 3,4-dioxygenase from Pseudomonas species strain CBS. J Biol Chem. 1986 Sep 25;261(27):12883–12888. [PubMed] [Google Scholar]
- Mondello F. J. Cloning and expression in Escherichia coli of Pseudomonas strain LB400 genes encoding polychlorinated biphenyl degradation. J Bacteriol. 1989 Mar;171(3):1725–1732. doi: 10.1128/jb.171.3.1725-1732.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perkins E. J., Gordon M. P., Caceres O., Lurquin P. F. Organization and sequence analysis of the 2,4-dichlorophenol hydroxylase and dichlorocatechol oxidative operons of plasmid pJP4. J Bacteriol. 1990 May;172(5):2351–2359. doi: 10.1128/jb.172.5.2351-2359.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reineke W., Knackmuss H. J. Microbial degradation of haloaromatics. Annu Rev Microbiol. 1988;42:263–287. doi: 10.1146/annurev.mi.42.100188.001403. [DOI] [PubMed] [Google Scholar]
- Schmidt E., Knackmuss H. J. Chemical structure and biodegradability of halogenated aromatic compounds. Conversion of chlorinated muconic acids into maleoylacetic acid. Biochem J. 1980 Oct 15;192(1):339–347. doi: 10.1042/bj1920339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt E., Remberg G., Knackmuss H. J. Chemical structure and biodegradability of halogenated aromatic compounds. Halogenated muconic acids as intermediates. Biochem J. 1980 Oct 15;192(1):331–337. doi: 10.1042/bj1920331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schraa G., Boone M. L., Jetten M. S., van Neerven A. R., Colberg P. J., Zehnder A. J. Degradation of 1,4-dichlorobenzene by Alcaligenes sp. strain A175. Appl Environ Microbiol. 1986 Dec;52(6):1374–1381. doi: 10.1128/aem.52.6.1374-1381.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweizer D., Markus A., Seez M., Ruf H. H., Lingens F. Purification and some properties of component B of the 4-chlorophenylacetate 3,4-dioxygenase from Pseudomonas species strain CBS 3. J Biol Chem. 1987 Jul 5;262(19):9340–9346. [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Spain J. C., Nishino S. F. Degradation of 1,4-dichlorobenzene by a Pseudomonas sp. Appl Environ Microbiol. 1987 May;53(5):1010–1019. doi: 10.1128/aem.53.5.1010-1019.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens G. M., Sidebotham J. M., Mann N. H., Dalton H. Cloning and expression in Escherichia coli of the toluene dioxygenase gene from Pseudomonas putida NCIB11767. FEMS Microbiol Lett. 1989 Feb;57(3):295–300. doi: 10.1016/0378-1097(89)90317-0. [DOI] [PubMed] [Google Scholar]
- Weisshaar M. P., Franklin F. C., Reineke W. Molecular cloning and expression of the 3-chlorobenzoate-degrading genes from Pseudomonas sp. strain B13. J Bacteriol. 1987 Jan;169(1):394–402. doi: 10.1128/jb.169.1.394-402.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyndham R. C., Singh R. K., Straus N. A. Catabolic instability, plasmid gene deletion and recombination in Alcaligenes sp. BR60. Arch Microbiol. 1988;150(3):237–243. doi: 10.1007/BF00407786. [DOI] [PubMed] [Google Scholar]
- Yanisch-Perron C., Vieira J., Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33(1):103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- Yen K. M., Serdar C. M. Genetics of naphthalene catabolism in pseudomonads. Crit Rev Microbiol. 1988;15(3):247–268. doi: 10.3109/10408418809104459. [DOI] [PubMed] [Google Scholar]
- Zylstra G. J., McCombie W. R., Gibson D. T., Finette B. A. Toluene degradation by Pseudomonas putida F1: genetic organization of the tod operon. Appl Environ Microbiol. 1988 Jun;54(6):1498–1503. doi: 10.1128/aem.54.6.1498-1503.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]



