Abstract
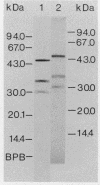
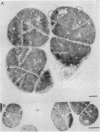
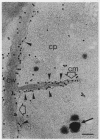
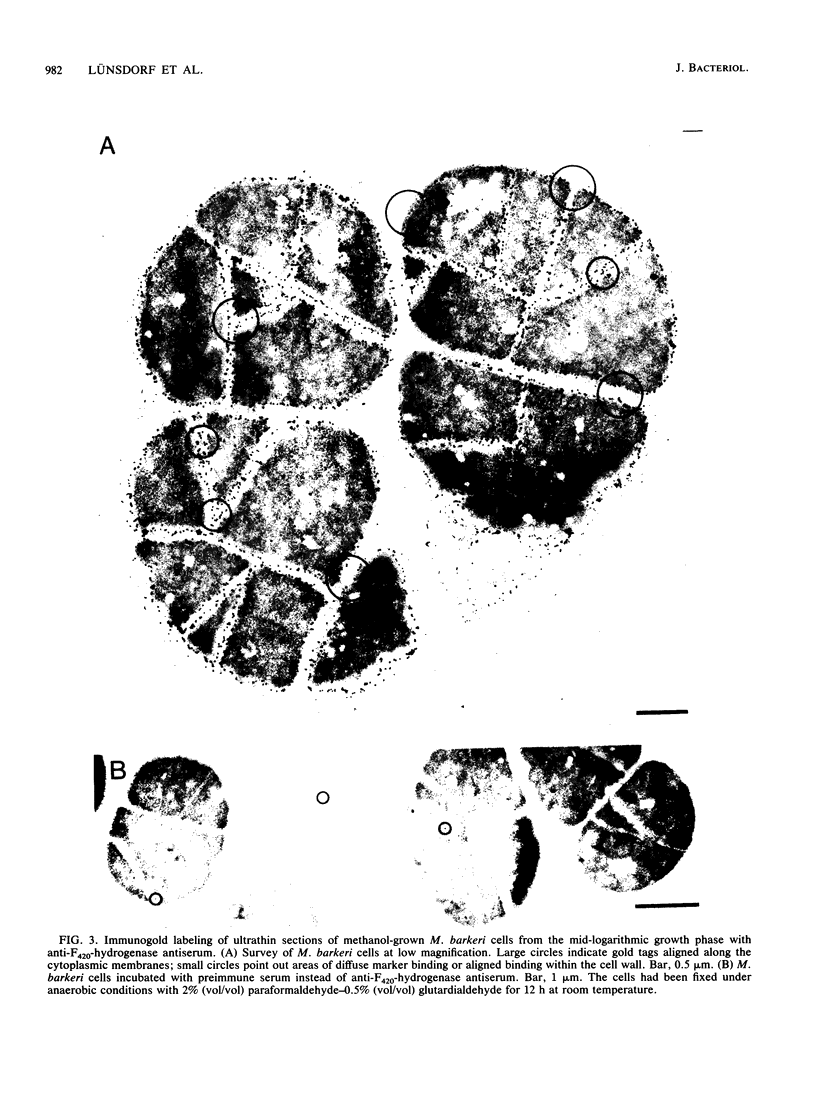
The cytological localization of the 8-hydroxy-5-deazaflavin (coenzyme F420)-reducing hydrogenase of Methanosarcina barkeri Fusaro was determined by immunoelectron microscopy, using a specific polyclonal rabbit antiserum raised against the homogeneous deazaflavin-dependent enzyme. In Western blot (immunoblot) experiments this antiserum reacted specifically with the native coenzyme F420-reducing hydrogenase, but did not cross-react with the coenzyme F420-nonreducing hydrogenase activity also detectable in crude extracts prepared from methanol-grown Methanosarcina cells. Immunogold labelling of ultrathin sections of anaerobically fixed methanol-grown cells from the exponential growth phase revealed that the coenzyme F420-reducing hydrogenase was predominantly located in the vicinity of the cytoplasmic membrane. From this result we concluded that the deazaflavin-dependent hydrogenase is associated with the cytoplasmic membrane in intact cells of M. barkeri during growth on methanol as the sole methanogenic substrate, and a possible role of this enzyme in the generation of the electrochemical proton gradient is discussed.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Batteiger B., Newhall W. J., 5th, Jones R. B. The use of Tween 20 as a blocking agent in the immunological detection of proteins transferred to nitrocellulose membranes. J Immunol Methods. 1982 Dec 30;55(3):297–307. doi: 10.1016/0022-1759(82)90089-8. [DOI] [PubMed] [Google Scholar]
- Blake M. S., Johnston K. H., Russell-Jones G. J., Gotschlich E. C. A rapid, sensitive method for detection of alkaline phosphatase-conjugated anti-antibody on Western blots. Anal Biochem. 1984 Jan;136(1):175–179. doi: 10.1016/0003-2697(84)90320-8. [DOI] [PubMed] [Google Scholar]
- Blaut M., Gottschalk G. Coupling of ATP synthesis and methane formation from methanol and molecular hydrogen in Methanosarcina barkeri. Eur J Biochem. 1984 May 15;141(1):217–222. doi: 10.1111/j.1432-1033.1984.tb08178.x. [DOI] [PubMed] [Google Scholar]
- Blaut M., Müller V., Fiebig K., Gottschalk G. Sodium ions and an energized membrane required by Methanosarcina barkeri for the oxidation of methanol to the level of formaldehyde. J Bacteriol. 1985 Oct;164(1):95–101. doi: 10.1128/jb.164.1.95-101.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976 May 7;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Daniels L., Sparling R., Sprott G. D. The bioenergetics of methanogenesis. Biochim Biophys Acta. 1984 Sep 6;768(2):113–163. doi: 10.1016/0304-4173(84)90002-8. [DOI] [PubMed] [Google Scholar]
- Fiebig K., Friedrich B. Purification of the F420-reducing hydrogenase from Methanosarcina barkeri (strain Fusaro). Eur J Biochem. 1989 Sep 1;184(1):79–88. doi: 10.1111/j.1432-1033.1989.tb14992.x. [DOI] [PubMed] [Google Scholar]
- Hauska G. Elucidation of methanogenesis seems well on its way. Trends Biochem Sci. 1988 Jan;13(1):2–4. doi: 10.1016/0968-0004(88)90003-5. [DOI] [PubMed] [Google Scholar]
- Hippe H., Caspari D., Fiebig K., Gottschalk G. Utilization of trimethylamine and other N-methyl compounds for growth and methane formation by Methanosarcina barkeri. Proc Natl Acad Sci U S A. 1979 Jan;76(1):494–498. doi: 10.1073/pnas.76.1.494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobson F. S., Daniels L., Fox J. A., Walsh C. T., Orme-Johnson W. H. Purification and properties of an 8-hydroxy-5-deazaflavin-reducing hydrogenase from Methanobacterium thermoautotrophicum. J Biol Chem. 1982 Apr 10;257(7):3385–3388. [PubMed] [Google Scholar]
- Muth E., Mörschel E., Klein A. Purification and characterization of an 8-hydroxy-5-deazaflavin-reducing hydrogenase from the archaebacterium Methanococcus voltae. Eur J Biochem. 1987 Dec 15;169(3):571–577. doi: 10.1111/j.1432-1033.1987.tb13647.x. [DOI] [PubMed] [Google Scholar]
- Noll K. M., Rinehart K. L., Jr, Tanner R. S., Wolfe R. S. Structure of component B (7-mercaptoheptanoylthreonine phosphate) of the methylcoenzyme M methylreductase system of Methanobacterium thermoautotrophicum. Proc Natl Acad Sci U S A. 1986 Jun;83(12):4238–4242. doi: 10.1073/pnas.83.12.4238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ossmer R., Mund T., Hartzell P. L., Konheiser U., Kohring G. W., Klein A., Wolfe R. S., Gottschalk G., Mayer F. Immunocytochemical localization of component C of the methylreductase system in Methanococcus voltae and Methanobacterium thermoautotrophicum. Proc Natl Acad Sci U S A. 1986 Aug;83(16):5789–5792. doi: 10.1073/pnas.83.16.5789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohde M., Mayer F., Meyer O. Immunocytochemical localization of carbon monoxide oxidase in Pseudomonas carboxydovorans. The enzyme is attached to the inner aspect of the cytoplasmic membrane. J Biol Chem. 1984 Dec 10;259(23):14788–14792. [PubMed] [Google Scholar]
- Roth J., Bendayan M., Carlemalm E., Villiger W., Garavito M. Enhancement of structural preservation and immunocytochemical staining in low temperature embedded pancreatic tissue. J Histochem Cytochem. 1981 May;29(5):663–671. doi: 10.1177/29.5.6166664. [DOI] [PubMed] [Google Scholar]
- Rouvière P. E., Bobik T. A., Wolfe R. S. Reductive activation of the methyl coenzyme M methylreductase system of Methanobacterium thermoautotrophicum delta H. J Bacteriol. 1988 Sep;170(9):3946–3952. doi: 10.1128/jb.170.9.3946-3952.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouvière P. E., Wolfe R. S. Component A3 of the methylcoenzyme M methylreductase system of Methanobacterium thermoautotrophicum delta H: resolution into two components. J Bacteriol. 1989 Sep;171(9):4556–4562. doi: 10.1128/jb.171.9.4556-4562.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouvière P. E., Wolfe R. S. Novel biochemistry of methanogenesis. J Biol Chem. 1988 Jun 15;263(17):7913–7916. [PubMed] [Google Scholar]
- Shapiro S., Wolfe R. S. Methyl-coenzyme M, an intermediate in methanogenic dissimilation of C1 compounds by Methanosarcina barkeri. J Bacteriol. 1980 Feb;141(2):728–734. doi: 10.1128/jb.141.2.728-734.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Towbin H., Staehelin T., Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci U S A. 1979 Sep;76(9):4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Meijden P., Heythuysen H. J., Pouwels A., Houwen F., van der Drift C., Vogels G. D. Methyltransferases involved in methanol conversion by Methanosarcina barkeri. Arch Microbiol. 1983 Jun;134(3):238–242. doi: 10.1007/BF00407765. [DOI] [PubMed] [Google Scholar]