Abstract
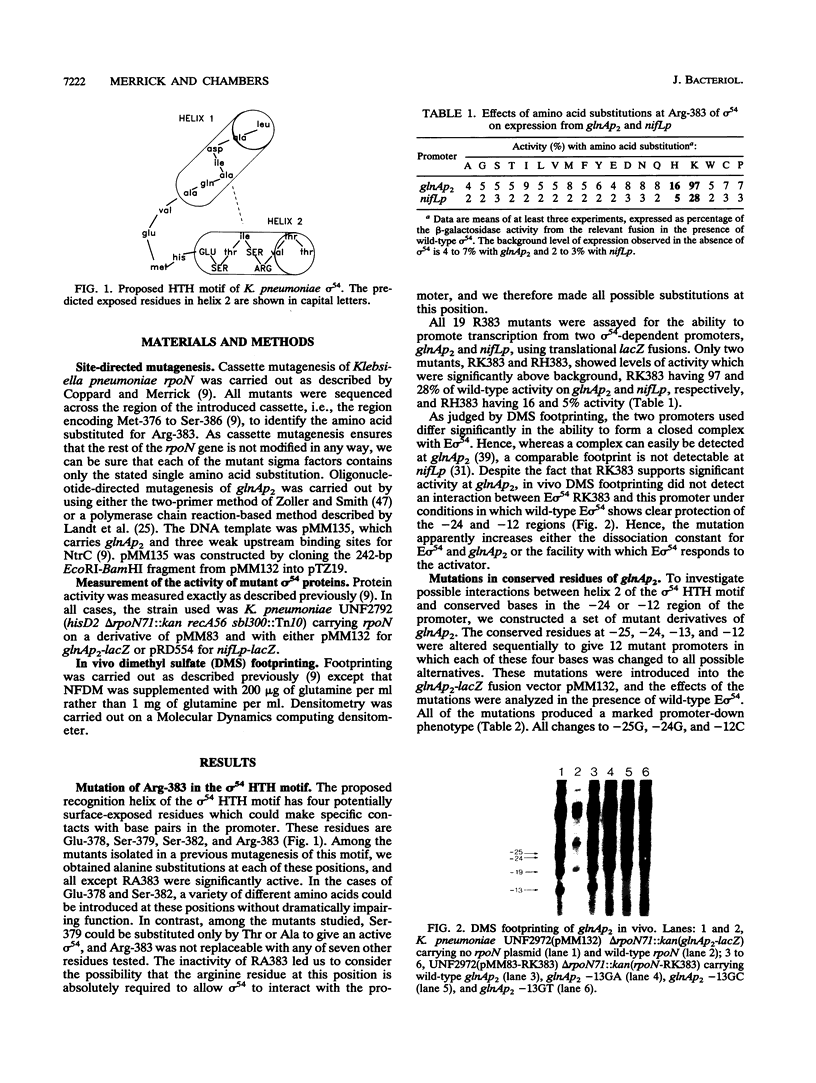
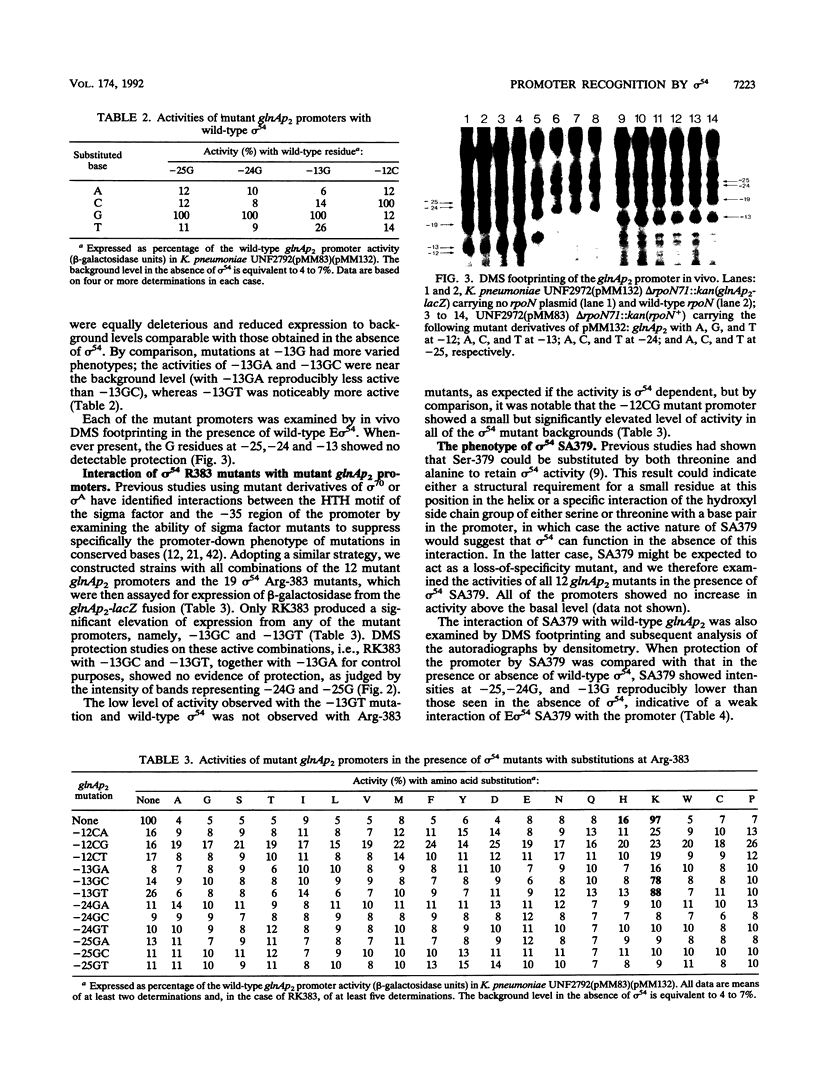
Residue Arg-383 in the proposed helix-turn-helix motif of the novel RNA polymerase sigma factor sigma 54 has been changed by site-directed mutagenesis to all possible alternative amino acids. Only two mutants, RK383 and RH383, are active in promoting transcription from either the glnAp2 promoter or the nifL promoter. We constructed a set of mutant derivatives of glnAp2 such that each base in the conserved GG and GC doublets at -24 and -12 was changed to all possible alternatives. All 12 mutant glnAp2 promoters showed a marked promoter-down phenotype with wild-type sigma 54, but RK383 suppressed changes of both G to C and G to T at -13. This result suggests that the sigma 54 helix-turn-helix is involved in recognition of the -13 region of sigma 54-dependent promoters.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Abel T., Maniatis T. Gene regulation. Action of leucine zippers. Nature. 1989 Sep 7;341(6237):24–25. doi: 10.1038/341024a0. [DOI] [PubMed] [Google Scholar]
- Aguilar O. M., Reiländer H., Arnold W., Pühler A. Rhizobium meliloti nifN (fixF) gene is part of an operon regulated by a nifA-dependent promoter and codes for a polypeptide homologous to the nifK gene product. J Bacteriol. 1987 Dec;169(12):5393–5400. doi: 10.1128/jb.169.12.5393-5400.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alias A., Cejudo F. J., Chabert J., Willison J. C., Vignais P. M. Nucleotide sequence of wild-type and mutant nifR4 (ntrA) genes of Rhodobacter capsulatus: identification of an essential glycine residue. Nucleic Acids Res. 1989 Jul 11;17(13):5377–5377. doi: 10.1093/nar/17.13.5377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Austin S., Dixon R. The prokaryotic enhancer binding protein NTRC has an ATPase activity which is phosphorylation and DNA dependent. EMBO J. 1992 Jun;11(6):2219–2228. doi: 10.1002/j.1460-2075.1992.tb05281.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beynon J., Cannon M., Buchanan-Wollaston V., Cannon F. The nif promoters of Klebsiella pneumoniae have a characteristic primary structure. Cell. 1983 Sep;34(2):665–671. doi: 10.1016/0092-8674(83)90399-9. [DOI] [PubMed] [Google Scholar]
- Buck M., Cannon W. Activator-independent formation of a closed complex between sigma 54-holoenzyme and nifH and nifU promoters of Klebsiella pneumoniae. Mol Microbiol. 1992 Jun;6(12):1625–1630. doi: 10.1111/j.1365-2958.1992.tb00887.x. [DOI] [PubMed] [Google Scholar]
- Buck M., Cannon W. Specific binding of the transcription factor sigma-54 to promoter DNA. Nature. 1992 Jul 30;358(6385):422–424. doi: 10.1038/358422a0. [DOI] [PubMed] [Google Scholar]
- Chan B., Busby S. Recognition of nucleotide sequences at the Escherichia coli galactose operon P1 promoter by RNA polymerase. Gene. 1989 Dec 14;84(2):227–236. doi: 10.1016/0378-1119(89)90496-4. [DOI] [PubMed] [Google Scholar]
- Coppard J. R., Merrick M. J. Cassette mutagenesis implicates a helix-turn-helix motif in promoter recognition by the novel RNA polymerase sigma factor sigma 54. Mol Microbiol. 1991 Jun;5(6):1309–1317. doi: 10.1111/j.1365-2958.1991.tb00777.x. [DOI] [PubMed] [Google Scholar]
- Daniels D., Zuber P., Losick R. Two amino acids in an RNA polymerase sigma factor involved in the recognition of adjacent base pairs in the -10 region of a cognate promoter. Proc Natl Acad Sci U S A. 1990 Oct;87(20):8075–8079. doi: 10.1073/pnas.87.20.8075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardella T., Moyle H., Susskind M. M. A mutant Escherichia coli sigma 70 subunit of RNA polymerase with altered promoter specificity. J Mol Biol. 1989 Apr 20;206(4):579–590. doi: 10.1016/0022-2836(89)90567-6. [DOI] [PubMed] [Google Scholar]
- Harrison S. C. A structural taxonomy of DNA-binding domains. Nature. 1991 Oct 24;353(6346):715–719. doi: 10.1038/353715a0. [DOI] [PubMed] [Google Scholar]
- Harrison S. C., Aggarwal A. K. DNA recognition by proteins with the helix-turn-helix motif. Annu Rev Biochem. 1990;59:933–969. doi: 10.1146/annurev.bi.59.070190.004441. [DOI] [PubMed] [Google Scholar]
- Helmann J. D., Chamberlin M. J. Structure and function of bacterial sigma factors. Annu Rev Biochem. 1988;57:839–872. doi: 10.1146/annurev.bi.57.070188.004203. [DOI] [PubMed] [Google Scholar]
- Hirschman J., Wong P. K., Sei K., Keener J., Kustu S. Products of nitrogen regulatory genes ntrA and ntrC of enteric bacteria activate glnA transcription in vitro: evidence that the ntrA product is a sigma factor. Proc Natl Acad Sci U S A. 1985 Nov;82(22):7525–7529. doi: 10.1073/pnas.82.22.7525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt T. P., Magasanik B. Transcription of glnA by purified Escherichia coli components: core RNA polymerase and the products of glnF, glnG, and glnL. Proc Natl Acad Sci U S A. 1985 Dec;82(24):8453–8457. doi: 10.1073/pnas.82.24.8453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones R., Haselkorn R. The DNA sequence of the Rhodobacter capsulatus ntrA, ntrB and ntrC gene analogues required for nitrogen fixation. Mol Gen Genet. 1989 Feb;215(3):507–516. doi: 10.1007/BF00427050. [DOI] [PubMed] [Google Scholar]
- Keilty S., Rosenberg M. Constitutive function of a positively regulated promoter reveals new sequences essential for activity. J Biol Chem. 1987 May 5;262(13):6389–6395. [PubMed] [Google Scholar]
- Kenney T. J., Moran C. P., Jr Genetic evidence for interaction of sigma A with two promoters in Bacillus subtilis. J Bacteriol. 1991 Jun;173(11):3282–3290. doi: 10.1128/jb.173.11.3282-3290.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kissinger C. R., Liu B. S., Martin-Blanco E., Kornberg T. B., Pabo C. O. Crystal structure of an engrailed homeodomain-DNA complex at 2.8 A resolution: a framework for understanding homeodomain-DNA interactions. Cell. 1990 Nov 2;63(3):579–590. doi: 10.1016/0092-8674(90)90453-l. [DOI] [PubMed] [Google Scholar]
- Kustu S., Santero E., Keener J., Popham D., Weiss D. Expression of sigma 54 (ntrA)-dependent genes is probably united by a common mechanism. Microbiol Rev. 1989 Sep;53(3):367–376. doi: 10.1128/mr.53.3.367-376.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landschulz W. H., Johnson P. F., McKnight S. L. The leucine zipper: a hypothetical structure common to a new class of DNA binding proteins. Science. 1988 Jun 24;240(4860):1759–1764. doi: 10.1126/science.3289117. [DOI] [PubMed] [Google Scholar]
- Landt O., Grunert H. P., Hahn U. A general method for rapid site-directed mutagenesis using the polymerase chain reaction. Gene. 1990 Nov 30;96(1):125–128. doi: 10.1016/0378-1119(90)90351-q. [DOI] [PubMed] [Google Scholar]
- Lonetto M., Gribskov M., Gross C. A. The sigma 70 family: sequence conservation and evolutionary relationships. J Bacteriol. 1992 Jun;174(12):3843–3849. doi: 10.1128/jb.174.12.3843-3849.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Losick R., Pero J. Cascades of Sigma factors. Cell. 1981 Sep;25(3):582–584. doi: 10.1016/0092-8674(81)90164-1. [DOI] [PubMed] [Google Scholar]
- Meijer W. G., Tabita F. R. Isolation and characterization of the nifUSVW-rpoN gene cluster from Rhodobacter sphaeroides. J Bacteriol. 1992 Jun;174(12):3855–3866. doi: 10.1128/jb.174.12.3855-3866.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merrick M. J., Gibbins J. R. The nucleotide sequence of the nitrogen-regulation gene ntrA of Klebsiella pneumoniae and comparison with conserved features in bacterial RNA polymerase sigma factors. Nucleic Acids Res. 1985 Nov 11;13(21):7607–7620. doi: 10.1093/nar/13.21.7607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merrick M., Gibbins J., Toukdarian A. The nucleotide sequence of the sigma factor gene ntrA (rpoN) of Azotobacter vinelandii: analysis of conserved sequences in NtrA proteins. Mol Gen Genet. 1987 Dec;210(2):323–330. doi: 10.1007/BF00325701. [DOI] [PubMed] [Google Scholar]
- Minchin S. D., Austin S., Dixon R. A. Transcriptional activation of the Klebsiella pneumoniae nifLA promoter by NTRC is face-of-the-helix dependent and the activator stabilizes the interaction of sigma 54-RNA polymerase with the promoter. EMBO J. 1989 Nov;8(11):3491–3499. doi: 10.1002/j.1460-2075.1989.tb08514.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morett E., Buck M. In vivo studies on the interaction of RNA polymerase-sigma 54 with the Klebsiella pneumoniae and Rhizobium meliloti nifH promoters. The role of NifA in the formation of an open promoter complex. J Mol Biol. 1989 Nov 5;210(1):65–77. doi: 10.1016/0022-2836(89)90291-x. [DOI] [PubMed] [Google Scholar]
- Nohno T., Saito T. Two transcriptional start sites found in the promoter region of Escherichia coli glutamine permease operon, glnHPQ. Nucleic Acids Res. 1987 Mar 25;15(6):2777–2777. doi: 10.1093/nar/15.6.2777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pabo C. O., Aggarwal A. K., Jordan S. R., Beamer L. J., Obeysekare U. R., Harrison S. C. Conserved residues make similar contacts in two repressor-operator complexes. Science. 1990 Mar 9;247(4947):1210–1213. doi: 10.1126/science.2315694. [DOI] [PubMed] [Google Scholar]
- Popham D. L., Szeto D., Keener J., Kustu S. Function of a bacterial activator protein that binds to transcriptional enhancers. Science. 1989 Feb 3;243(4891):629–635. doi: 10.1126/science.2563595. [DOI] [PubMed] [Google Scholar]
- Quinto C., De La Vega H., Flores M., Leemans J., Cevallos M. A., Pardo M. A., Azpiroz R., De Lourdes Girard M., Calva E., Palacios R. Nitrogenase reductase: A functional multigene family in Rhizobium phaseoli. Proc Natl Acad Sci U S A. 1985 Feb;82(4):1170–1174. doi: 10.1073/pnas.82.4.1170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reitzer L. J., Bueno R., Cheng W. D., Abrams S. A., Rothstein D. M., Hunt T. P., Tyler B., Magasanik B. Mutations that create new promoters suppress the sigma 54 dependence of glnA transcription in Escherichia coli. J Bacteriol. 1987 Sep;169(9):4279–4284. doi: 10.1128/jb.169.9.4279-4284.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reznikoff W. S., Siegele D. A., Cowing D. W., Gross C. A. The regulation of transcription initiation in bacteria. Annu Rev Genet. 1985;19:355–387. doi: 10.1146/annurev.ge.19.120185.002035. [DOI] [PubMed] [Google Scholar]
- Sasse-Dwight S., Gralla J. D. Probing the Escherichia coli glnALG upstream activation mechanism in vivo. Proc Natl Acad Sci U S A. 1988 Dec;85(23):8934–8938. doi: 10.1073/pnas.85.23.8934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasse-Dwight S., Gralla J. D. Role of eukaryotic-type functional domains found in the prokaryotic enhancer receptor factor sigma 54. Cell. 1990 Sep 7;62(5):945–954. doi: 10.1016/0092-8674(90)90269-k. [DOI] [PubMed] [Google Scholar]
- Scott K. F., Rolfe B. G., Shine J. Biological nitrogen fixation: primary structure of the Rhizobium trifolii iron protein gene. DNA. 1983;2(2):149–155. doi: 10.1089/dna.1983.2.149. [DOI] [PubMed] [Google Scholar]
- Siegele D. A., Hu J. C., Walter W. A., Gross C. A. Altered promoter recognition by mutant forms of the sigma 70 subunit of Escherichia coli RNA polymerase. J Mol Biol. 1989 Apr 20;206(4):591–603. doi: 10.1016/0022-2836(89)90568-8. [DOI] [PubMed] [Google Scholar]
- Vinson C. R., Sigler P. B., McKnight S. L. Scissors-grip model for DNA recognition by a family of leucine zipper proteins. Science. 1989 Nov 17;246(4932):911–916. doi: 10.1126/science.2683088. [DOI] [PubMed] [Google Scholar]
- Waldburger C., Gardella T., Wong R., Susskind M. M. Changes in conserved region 2 of Escherichia coli sigma 70 affecting promoter recognition. J Mol Biol. 1990 Sep 20;215(2):267–276. doi: 10.1016/s0022-2836(05)80345-6. [DOI] [PubMed] [Google Scholar]
- Weiner L., Brissette J. L., Model P. Stress-induced expression of the Escherichia coli phage shock protein operon is dependent on sigma 54 and modulated by positive and negative feedback mechanisms. Genes Dev. 1991 Oct;5(10):1912–1923. doi: 10.1101/gad.5.10.1912. [DOI] [PubMed] [Google Scholar]
- Weiss D. S., Batut J., Klose K. E., Keener J., Kustu S. The phosphorylated form of the enhancer-binding protein NTRC has an ATPase activity that is essential for activation of transcription. Cell. 1991 Oct 4;67(1):155–167. doi: 10.1016/0092-8674(91)90579-n. [DOI] [PubMed] [Google Scholar]
- Zoller M. J., Smith M. Oligonucleotide-directed mutagenesis: a simple method using two oligonucleotide primers and a single-stranded DNA template. DNA. 1984 Dec;3(6):479–488. doi: 10.1089/dna.1.1984.3.479. [DOI] [PubMed] [Google Scholar]
- Zuber P., Healy J., Carter H. L., 3rd, Cutting S., Moran C. P., Jr, Losick R. Mutation changing the specificity of an RNA polymerase sigma factor. J Mol Biol. 1989 Apr 20;206(4):605–614. doi: 10.1016/0022-2836(89)90569-x. [DOI] [PubMed] [Google Scholar]




