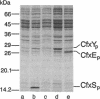
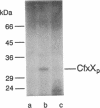
Abstract
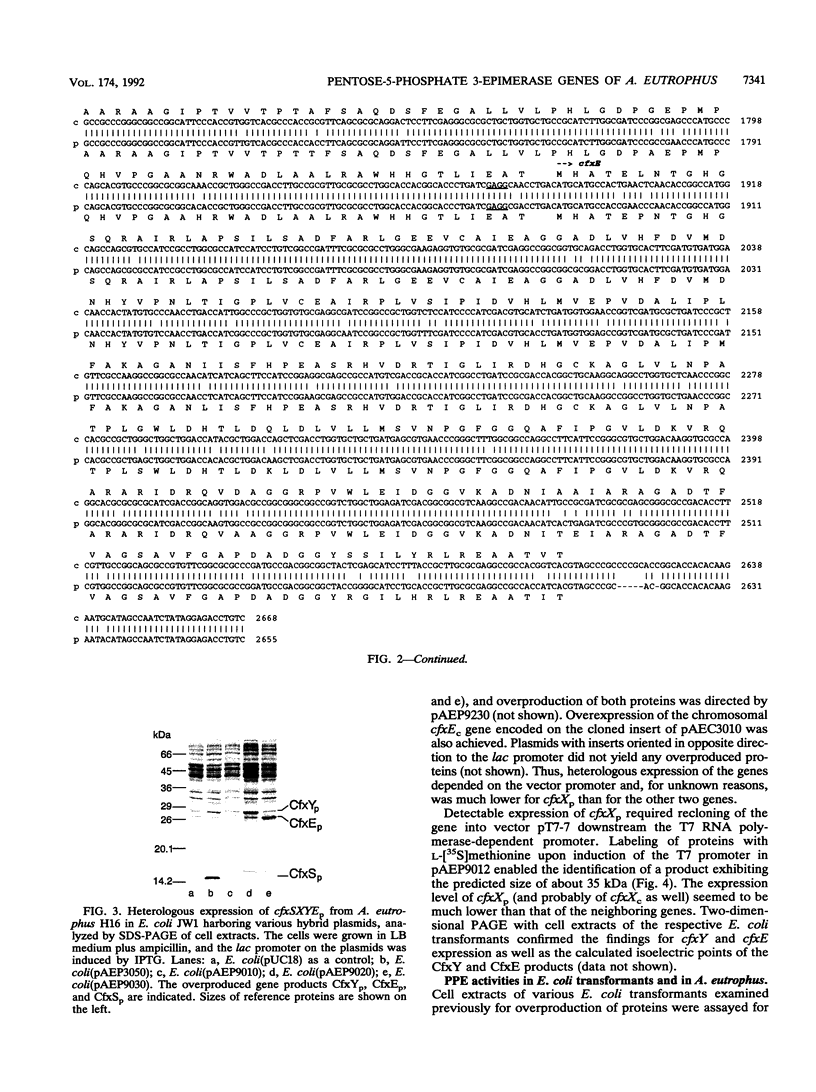
Several genes (cfx genes) encoding Calvin cycle enzymes in Alcaligenes eutrophus are organized in two highly homologous operons comprising at least 11 kb. One cfx operon is located on the chromosome; the other is located on megaplasmid pHG1 of the organism (B. Bowien, U. Windhövel, J.-G. Yoo, R. Bednarski, and B. Kusian, FEMS Microbiol. Rev. 87:445-450, 1990). Corresponding regions of about 2.7 kb from within the operons were sequenced. Three open reading frames, designated cfxX (954 bp), cfxY (765 bp), and cfxE (726 bp), were detected at equivalent positions in the two sequences. The nucleotide identity of the sequences amounted to 94%. Heterologous expression of the subcloned pHG1-encoded open reading frames in Escherichia coli suggested that they were functional genes. The observed sizes of the gene products CfxX (35 kDa), CfxY (27 kDa), and CfxE (25.5 kDa) closely corresponded to the values calculated on the basis of the sequence information. E. coli clones harboring the cfxE gene showed up to about 19-fold-higher activities of pentose-5-phosphate 3-epimerase (PPE; EC 5.1.3.1) than did reference clones, suggesting that cfxE encodes PPE, another Calvin cycle enzyme. These data agree with the finding that in A. eutrophus, PPE activity is significantly enhanced under autotrophic growth conditions which lead to a derepression of the cfx operons. No functions could be assigned to CfxX and CfxY.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Assali N. E., Martin W. F., Sommerville C. C., Loiseaux-de Goër S. Evolution of the Rubisco operon from prokaryotes to algae: structure and analysis of the rbcS gene of the brown alga Pylaiella littoralis. Plant Mol Biol. 1991 Oct;17(4):853–863. doi: 10.1007/BF00037066. [DOI] [PubMed] [Google Scholar]
- Birnboim H. C., Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979 Nov 24;7(6):1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boczar B. A., Delaney T. P., Cattolico R. A. Gene for the ribulose-1,5-bisphosphate carboxylase small subunit protein of the marine chromophyte Olisthodiscus luteus is similar to that of a chemoautotrophic bacterium. Proc Natl Acad Sci U S A. 1989 Jul;86(13):4996–4999. doi: 10.1073/pnas.86.13.4996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowien B., Schlegel H. G. Der Biosyntheseweg der RNS-Ribose in Hydrogenomonas eutropha Stamm H 16 und Pseudomonas facilis. Arch Mikrobiol. 1972;85(2):95–112. doi: 10.1007/BF00409291. [DOI] [PubMed] [Google Scholar]
- Bowien B., Schlegel H. G. Physiology and biochemistry of aerobic hydrogen-oxidizing bacteria. Annu Rev Microbiol. 1981;35:405–452. doi: 10.1146/annurev.mi.35.100181.002201. [DOI] [PubMed] [Google Scholar]
- Foulger D., Errington J. Sequential activation of dual promoters by different sigma factors maintains spoVJ expression during successive developmental stages of Bacillus subtilis. Mol Microbiol. 1991 Jun;5(6):1363–1373. doi: 10.1111/j.1365-2958.1991.tb00783.x. [DOI] [PubMed] [Google Scholar]
- Friedrich B., Hogrefe C., Schlegel H. G. Naturally occurring genetic transfer of hydrogen-oxidizing ability between strains of Alcaligenes eutrophus. J Bacteriol. 1981 Jul;147(1):198–205. doi: 10.1128/jb.147.1.198-205.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedrich C. G. Depression of hydrogenase during limitation of electron donors and derepression of ribulosebisphosphate carboxylase during carbon limitation of Alcaligenes eutrophus. J Bacteriol. 1982 Jan;149(1):203–210. doi: 10.1128/jb.149.1.203-210.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson J. L., Falcone D. L., Tabita F. R. Nucleotide sequence, transcriptional analysis, and expression of genes encoded within the form I CO2 fixation operon of Rhodobacter sphaeroides. J Biol Chem. 1991 Aug 5;266(22):14646–14653. [PubMed] [Google Scholar]
- Hanahan D. Studies on transformation of Escherichia coli with plasmids. J Mol Biol. 1983 Jun 5;166(4):557–580. doi: 10.1016/s0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- Higgins C. F., Hiles I. D., Salmond G. P., Gill D. R., Downie J. A., Evans I. J., Holland I. B., Gray L., Buckel S. D., Bell A. W. A family of related ATP-binding subunits coupled to many distinct biological processes in bacteria. Nature. 1986 Oct 2;323(6087):448–450. doi: 10.1038/323448a0. [DOI] [PubMed] [Google Scholar]
- Holmes D. S., Quigley M. A rapid boiling method for the preparation of bacterial plasmids. Anal Biochem. 1981 Jun;114(1):193–197. doi: 10.1016/0003-2697(81)90473-5. [DOI] [PubMed] [Google Scholar]
- Horiuchi K., Zinder N. D. Azure mutants: a type of host-dependent mutant of the bacteriophage f2. Science. 1967 Jun 23;156(3782):1618–1623. doi: 10.1126/science.156.3782.1618. [DOI] [PubMed] [Google Scholar]
- Hwang S. R., Tabita F. R. Cotranscription, deduced primary structure, and expression of the chloroplast-encoded rbcL and rbcS genes of the marine diatom Cylindrotheca sp. strain N1. J Biol Chem. 1991 Apr 5;266(10):6271–6279. [PubMed] [Google Scholar]
- Jendrossek D., Steinbüchel A., Schlegel H. G. Alcohol dehydrogenase gene from Alcaligenes eutrophus: subcloning, heterologous expression in Escherichia coli, sequencing, and location of Tn5 insertions. J Bacteriol. 1988 Nov;170(11):5248–5256. doi: 10.1128/jb.170.11.5248-5256.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karmali A., Drake A. F., Spencer N. Purification, properties and assay of D-ribulose 5-phosphate 3-epimerase from human erythrocytes. Biochem J. 1983 Jun 1;211(3):617–623. doi: 10.1042/bj2110617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kossmann J., Klintworth R., Bowien B. Sequence analysis of the chromosomal and plasmid genes encoding phosphoribulokinase from Alcaligenes eutrophus. Gene. 1989 Dec 21;85(1):247–252. doi: 10.1016/0378-1119(89)90490-3. [DOI] [PubMed] [Google Scholar]
- Kostrzewa M., Valentin K., Maid U., Radetzky R., Zetsche K. Structure of the rubisco operon from the multicellular red alga Antithamnion spec. Curr Genet. 1990 Dec;18(5):465–469. doi: 10.1007/BF00309918. [DOI] [PubMed] [Google Scholar]
- LOWRY O. H., ROSEBROUGH N. J., FARR A. L., RANDALL R. J. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951 Nov;193(1):265–275. [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lee N., Gielow W., Martin R., Hamilton E., Fowler A. The organization of the araBAD operon of Escherichia coli. Gene. 1986;47(2-3):231–244. doi: 10.1016/0378-1119(86)90067-3. [DOI] [PubMed] [Google Scholar]
- Lin H. C., Lei S. P., Studnicka G., Wilcox G. The araBAD operon of Salmonella typhimurium LT2. III. Nucleotide sequence of araD and its flanking regions, and primary structure of its product, L-ribulose-5-phosphate 4-epimerase. Gene. 1985;34(1):129–134. doi: 10.1016/0378-1119(85)90303-8. [DOI] [PubMed] [Google Scholar]
- Mandel M., Higa A. Calcium-dependent bacteriophage DNA infection. J Mol Biol. 1970 Oct 14;53(1):159–162. doi: 10.1016/0022-2836(70)90051-3. [DOI] [PubMed] [Google Scholar]
- Meijer W. G., Arnberg A. C., Enequist H. G., Terpstra P., Lidstrom M. E., Dijkhuizen L. Identification and organization of carbon dioxide fixation genes in Xanthobacter flavus H4-14. Mol Gen Genet. 1991 Feb;225(2):320–330. doi: 10.1007/BF00269865. [DOI] [PubMed] [Google Scholar]
- O'Farrell P. H. High resolution two-dimensional electrophoresis of proteins. J Biol Chem. 1975 May 25;250(10):4007–4021. [PMC free article] [PubMed] [Google Scholar]
- Pearson W. R., Lipman D. J. Improved tools for biological sequence comparison. Proc Natl Acad Sci U S A. 1988 Apr;85(8):2444–2448. doi: 10.1073/pnas.85.8.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peoples O. P., Sinskey A. J. Poly-beta-hydroxybutyrate (PHB) biosynthesis in Alcaligenes eutrophus H16. Identification and characterization of the PHB polymerase gene (phbC). J Biol Chem. 1989 Sep 15;264(26):15298–15303. [PubMed] [Google Scholar]
- Peoples O. P., Sinskey A. J. Poly-beta-hydroxybutyrate biosynthesis in Alcaligenes eutrophus H16. Characterization of the genes encoding beta-ketothiolase and acetoacetyl-CoA reductase. J Biol Chem. 1989 Sep 15;264(26):15293–15297. [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shine J., Dalgarno L. Determinant of cistron specificity in bacterial ribosomes. Nature. 1975 Mar 6;254(5495):34–38. doi: 10.1038/254034a0. [DOI] [PubMed] [Google Scholar]
- Tabor S., Richardson C. C. A bacteriophage T7 RNA polymerase/promoter system for controlled exclusive expression of specific genes. Proc Natl Acad Sci U S A. 1985 Feb;82(4):1074–1078. doi: 10.1073/pnas.82.4.1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terada T., Mukae H., Ohashi K., Hosomi S., Mizoguchi T., Uehara K. Characterization of an enzyme which catalyzes isomerization and epimerization of D-erythrose 4-phosphate. Eur J Biochem. 1985 Apr 15;148(2):345–351. doi: 10.1111/j.1432-1033.1985.tb08845.x. [DOI] [PubMed] [Google Scholar]
- Vieira J., Messing J. The pUC plasmids, an M13mp7-derived system for insertion mutagenesis and sequencing with synthetic universal primers. Gene. 1982 Oct;19(3):259–268. doi: 10.1016/0378-1119(82)90015-4. [DOI] [PubMed] [Google Scholar]
- Vogelstein B., Gillespie D. Preparative and analytical purification of DNA from agarose. Proc Natl Acad Sci U S A. 1979 Feb;76(2):615–619. doi: 10.1073/pnas.76.2.615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Windhövel U., Bowien B. Identification of cfxR, an activator gene of autotrophic CO2 fixation in Alcaligenes eutrophus. Mol Microbiol. 1991 Nov;5(11):2695–2705. doi: 10.1111/j.1365-2958.1991.tb01978.x. [DOI] [PubMed] [Google Scholar]
- Windhövel U., Bowien B. On the operon structure of the cfx gene clusters in Alcaligenes eutrophus. Arch Microbiol. 1990;154(1):85–91. doi: 10.1007/BF00249183. [DOI] [PubMed] [Google Scholar]
- Wood T. Purification and properties of D-ribulose-5-phosphate 3-epimerase from calf liver. Biochim Biophys Acta. 1979 Oct 11;570(2):352–362. doi: 10.1016/0005-2744(79)90155-4. [DOI] [PubMed] [Google Scholar]
- Yanisch-Perron C., Vieira J., Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33(1):103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]