Abstract
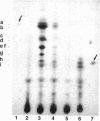
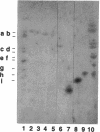
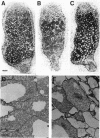
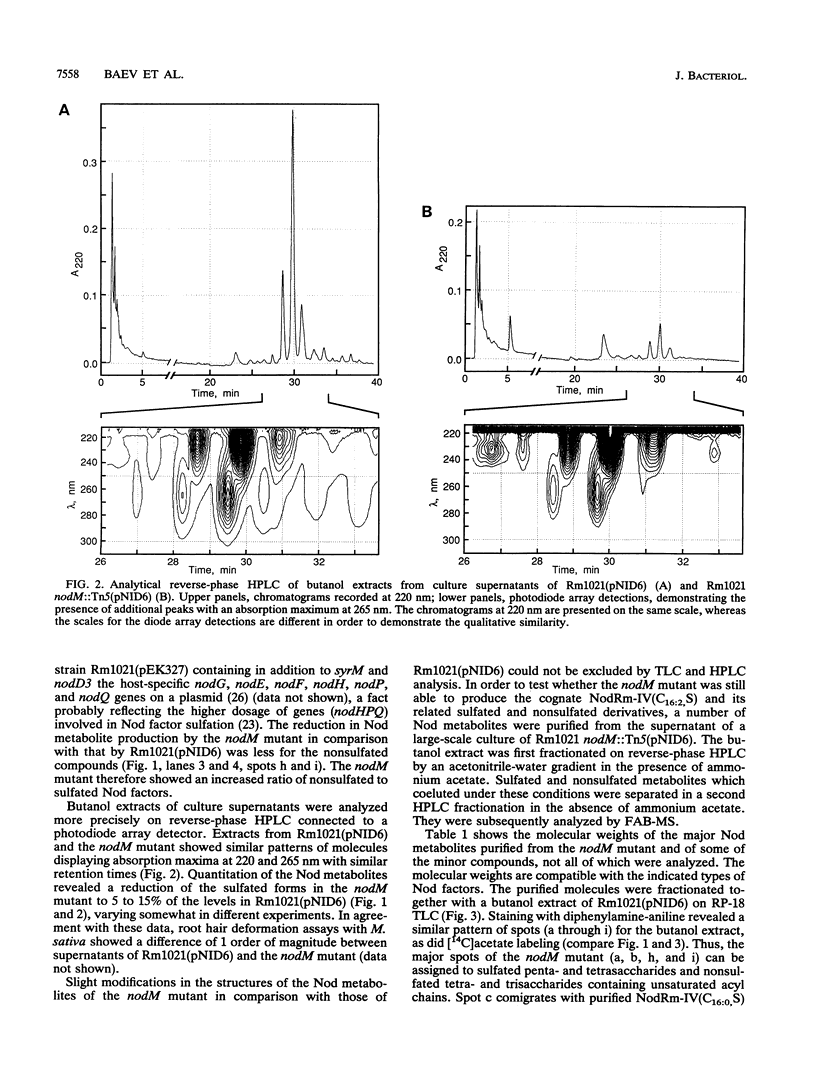
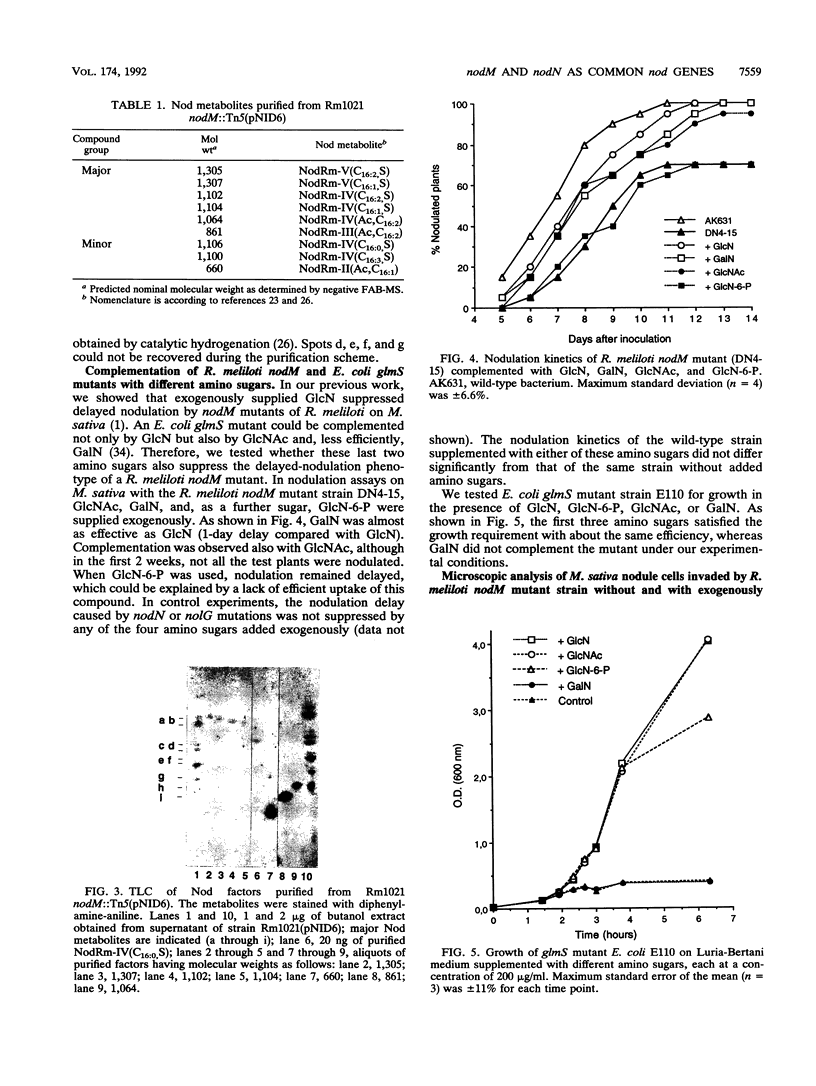
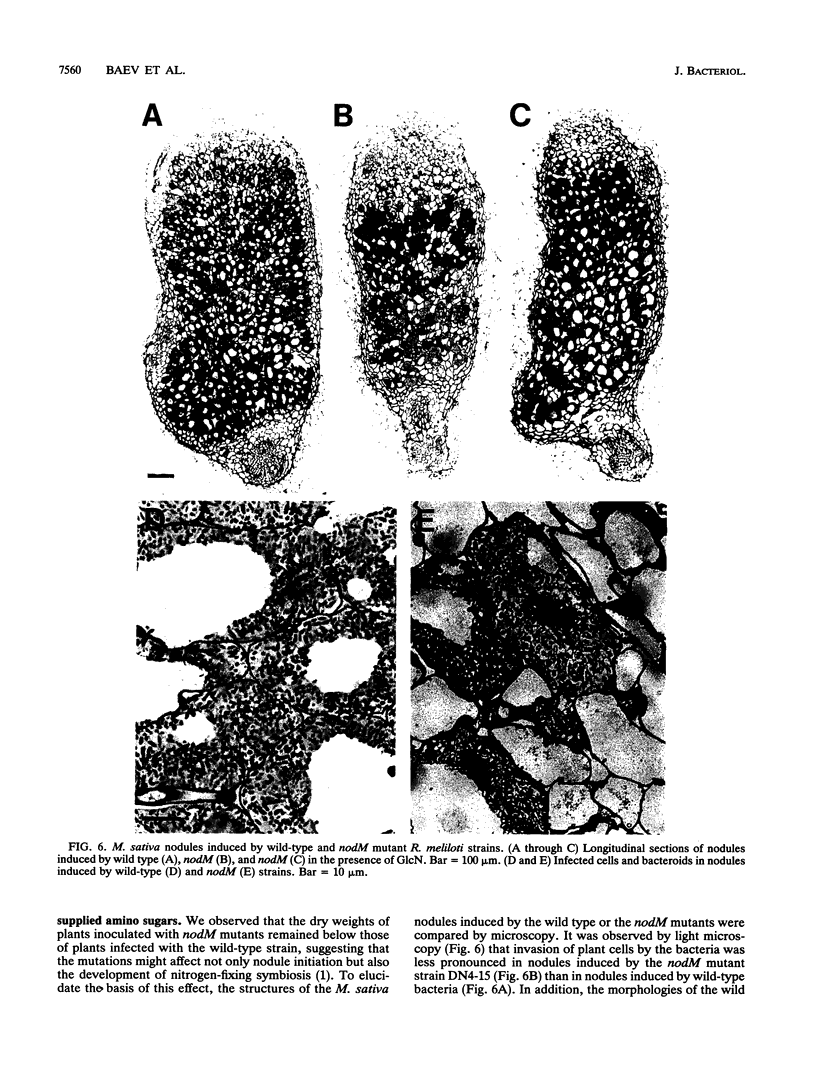
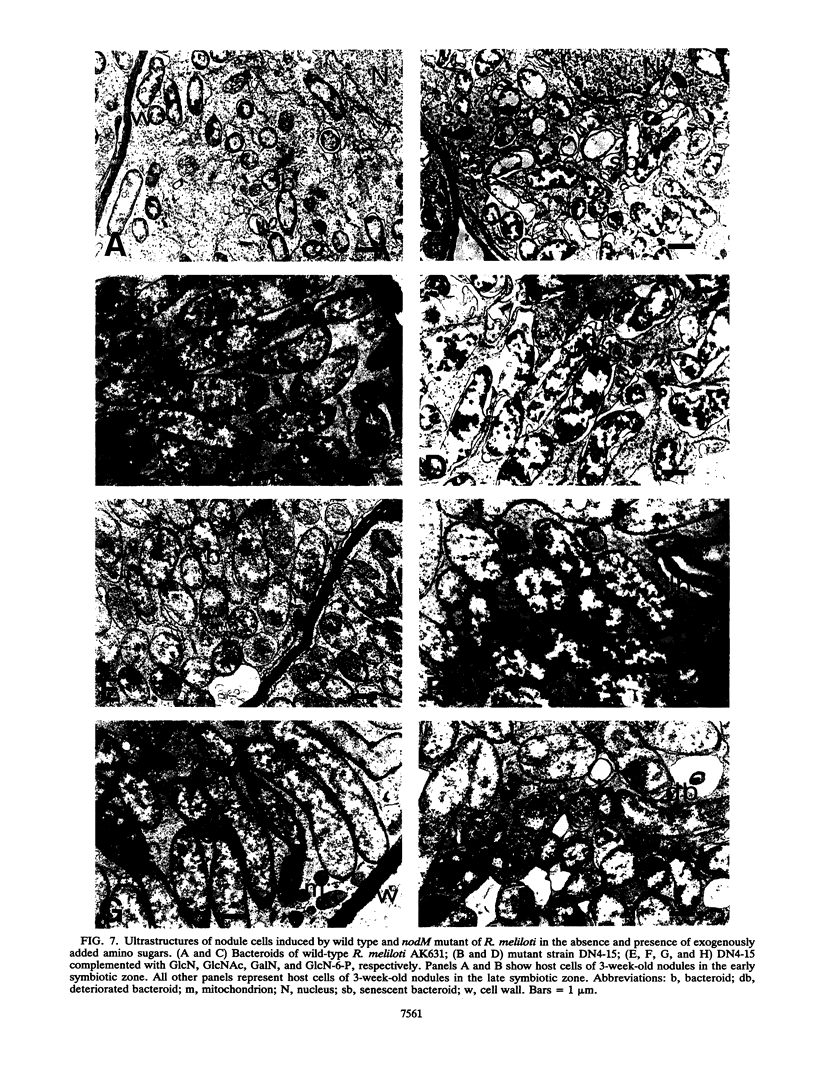
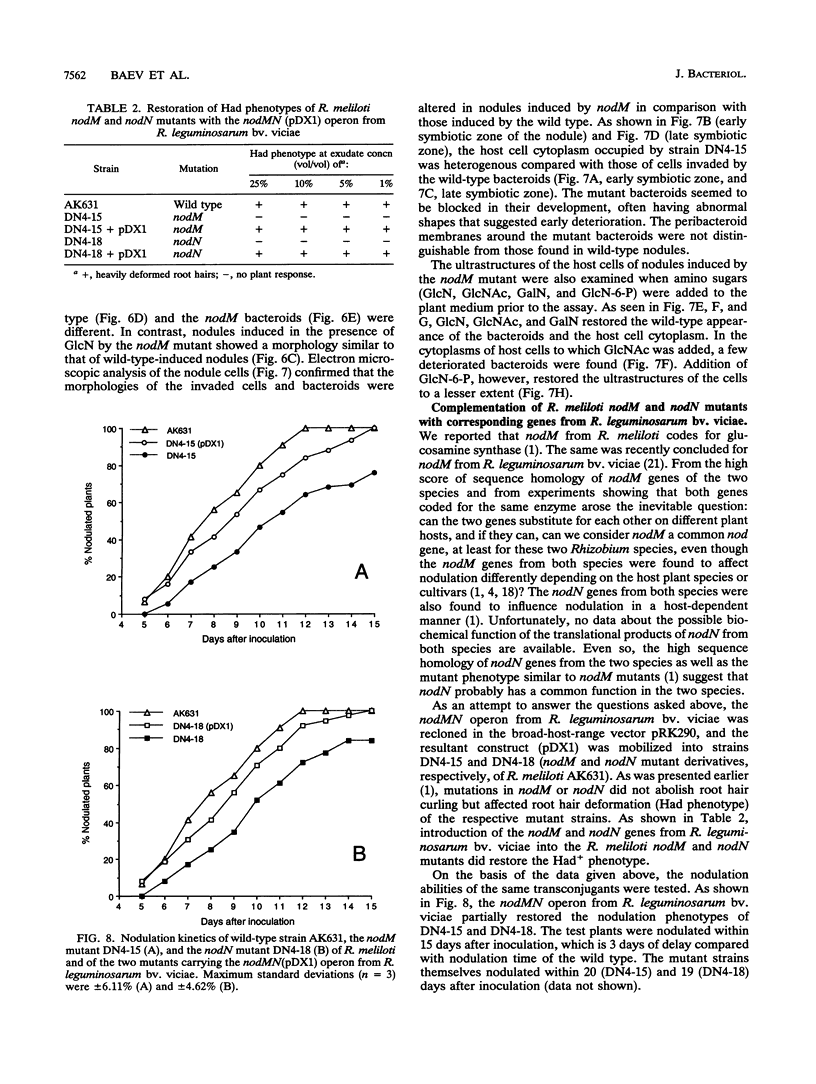
Earlier, we showed that Rhizobium meliloti nodM codes for glucosamine synthase and that nodM and nodN mutants produce strongly reduced root hair deformation activity and display delayed nodulation of Medicago sativa (Baev et al., Mol. Gen. Genet. 228:113-124, 1991). Here, we demonstrate that nodM and nodN genes from Rhizobium leguminosarum biovar viciae restore the root hair deformation activity of exudates of the corresponding R. meliloti mutant strains. Partial restoration of the nodulation phenotypes of these two strains was also observed. In nodulation assays, galactosamine and N-acetylglucosamine could substitute for glucosamine in the suppression of the R. meliloti nodM mutation, although N-acetylglucosamine was less efficient. We observed that in nodules induced by nodM mutants, the bacteroids did not show complete development or were deteriorated, resulting in decreased nitrogen fixation and, consequently, lower dry weights of the plants. This mutant phenotype could also be suppressed by exogenously supplied glucosamine, N-acetylglucosamine, and galactosamine and to a lesser extent by glucosamine-6-phosphate, indicating that the nodM mutant bacteroids are limited for glucosamine. In addition, by using derivatives of the wild type and a nodM mutant in which the nod genes are expressed at a high constitutive level, it was shown that the nodM mutant produces significantly fewer Nod factors than the wild-type strain but that their chemical structures are unchanged. However, the relative amounts of analogs of the cognate Nod signals were elevated, and this may explain the observed host range effects of the nodM mutation. Our data indicate that both the nodM and nodN genes of the two species have common functions and confirm that NodM is a glucosamine synthase with the biochemical role of providing sufficient amounts of the sugar moiety for the synthesis of the glucosamine oligosaccharide signal molecules.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Baev N., Endre G., Petrovics G., Banfalvi Z., Kondorosi A. Six nodulation genes of nod box locus 4 in Rhizobium meliloti are involved in nodulation signal production: nodM codes for D-glucosamine synthetase. Mol Gen Genet. 1991 Aug;228(1-2):113–124. doi: 10.1007/BF00282455. [DOI] [PubMed] [Google Scholar]
- Banfalvi Z., Kondorosi A. Production of root hair deformation factors by Rhizobium meliloti nodulation genes in Escherichia coli: HsnD (NodH) is involved in the plant host-specific modification of the NodABC factor. Plant Mol Biol. 1989 Jul;13(1):1–12. doi: 10.1007/BF00027330. [DOI] [PubMed] [Google Scholar]
- Canter Cremers H., Spaink H. P., Wijfjes A. H., Pees E., Wijffelman C. A., Okker R. J., Lugtenberg B. J. Additional nodulation genes on the Sym plasmid of Rhizobium leguminosarum biovar viciae. Plant Mol Biol. 1989 Aug;13(2):163–174. doi: 10.1007/BF00016135. [DOI] [PubMed] [Google Scholar]
- Chang A. C., Cohen S. N. Construction and characterization of amplifiable multicopy DNA cloning vehicles derived from the P15A cryptic miniplasmid. J Bacteriol. 1978 Jun;134(3):1141–1156. doi: 10.1128/jb.134.3.1141-1156.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ditta G., Stanfield S., Corbin D., Helinski D. R. Broad host range DNA cloning system for gram-negative bacteria: construction of a gene bank of Rhizobium meliloti. Proc Natl Acad Sci U S A. 1980 Dec;77(12):7347–7351. doi: 10.1073/pnas.77.12.7347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faucher C., Maillet F., Vasse J., Rosenberg C., van Brussel A. A., Truchet G., Dénarié J. Rhizobium meliloti host range nodH gene determines production of an alfalfa-specific extracellular signal. J Bacteriol. 1988 Dec;170(12):5489–5499. doi: 10.1128/jb.170.12.5489-5499.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirsch A. M., Long S. R., Bang M., Haskins N., Ausubel F. M. Structural studies of alfalfa roots infected with nodulation mutants of Rhizobium meliloti. J Bacteriol. 1982 Jul;151(1):411–419. doi: 10.1128/jb.151.1.411-419.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horvath B., Kondorosi E., John M., Schmidt J., Török I., Györgypal Z., Barabas I., Wieneke U., Schell J., Kondorosi A. Organization, structure and symbiotic function of Rhizobium meliloti nodulation genes determining host specificity for alfalfa. Cell. 1986 Aug 1;46(3):335–343. doi: 10.1016/0092-8674(86)90654-9. [DOI] [PubMed] [Google Scholar]
- Kondorosi A., Kondorosi E., John M., Schmidt J., Schell J. The role of nodulation genes in bacterium-plant communication. Genet Eng (N Y) 1991;13:115–136. doi: 10.1007/978-1-4615-3760-1_4. [DOI] [PubMed] [Google Scholar]
- Kondorosi E., Gyuris J., Schmidt J., John M., Duda E., Hoffmann B., Schell J., Kondorosi A. Positive and negative control of nod gene expression in Rhizobium meliloti is required for optimal nodulation. EMBO J. 1989 May;8(5):1331–1340. doi: 10.1002/j.1460-2075.1989.tb03513.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondorosi E., Pierre M., Cren M., Haumann U., Buiré M., Hoffmann B., Schell J., Kondorosi A. Identification of NolR, a negative transacting factor controlling the nod regulon in Rhizobium meliloti. J Mol Biol. 1991 Dec 20;222(4):885–896. doi: 10.1016/0022-2836(91)90583-r. [DOI] [PubMed] [Google Scholar]
- Lerouge P., Roche P., Faucher C., Maillet F., Truchet G., Promé J. C., Dénarié J. Symbiotic host-specificity of Rhizobium meliloti is determined by a sulphated and acylated glucosamine oligosaccharide signal. Nature. 1990 Apr 19;344(6268):781–784. doi: 10.1038/344781a0. [DOI] [PubMed] [Google Scholar]
- Lewis-Henderson W. R., Djordjevic M. A. A cultivar-specific interaction between Rhizobium leguminosarum bv. trifolii and subterranean clover is controlled by nodM, other bacterial cultivar specificity genes, and a single recessive host gene. J Bacteriol. 1991 May;173(9):2791–2799. doi: 10.1128/jb.173.9.2791-2799.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long S. R. Rhizobium-legume nodulation: life together in the underground. Cell. 1989 Jan 27;56(2):203–214. doi: 10.1016/0092-8674(89)90893-3. [DOI] [PubMed] [Google Scholar]
- Marie C., Barny M. A., Downie J. A. Rhizobium leguminosarum has two glucosamine synthases, GlmS and NodM, required for nodulation and development of nitrogen-fixing nodules. Mol Microbiol. 1992 Apr;6(7):843–851. doi: 10.1111/j.1365-2958.1992.tb01535.x. [DOI] [PubMed] [Google Scholar]
- REYNOLDS E. S. The use of lead citrate at high pH as an electron-opaque stain in electron microscopy. J Cell Biol. 1963 Apr;17:208–212. doi: 10.1083/jcb.17.1.208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roche P., Debellé F., Maillet F., Lerouge P., Faucher C., Truchet G., Dénarié J., Promé J. C. Molecular basis of symbiotic host specificity in Rhizobium meliloti: nodH and nodPQ genes encode the sulfation of lipo-oligosaccharide signals. Cell. 1991 Dec 20;67(6):1131–1143. doi: 10.1016/0092-8674(91)90290-f. [DOI] [PubMed] [Google Scholar]
- Rostas K., Kondorosi E., Horvath B., Simoncsits A., Kondorosi A. Conservation of extended promoter regions of nodulation genes in Rhizobium. Proc Natl Acad Sci U S A. 1986 Mar;83(6):1757–1761. doi: 10.1073/pnas.83.6.1757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlaman H. R., Horvath B., Vijgenboom E., Okker R. J., Lugtenberg B. J. Suppression of nodulation gene expression in bacteroids of Rhizobium leguminosarum biovar viciae. J Bacteriol. 1991 Jul;173(14):4277–4287. doi: 10.1128/jb.173.14.4277-4287.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt J., Wingender R., John M., Wieneke U., Schell J. Rhizobium meliloti nodA and nodB genes are involved in generating compounds that stimulate mitosis of plant cells. Proc Natl Acad Sci U S A. 1988 Nov;85(22):8578–8582. doi: 10.1073/pnas.85.22.8578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultze M., Quiclet-Sire B., Kondorosi E., Virelizer H., Glushka J. N., Endre G., Géro S. D., Kondorosi A. Rhizobium meliloti produces a family of sulfated lipooligosaccharides exhibiting different degrees of plant host specificity. Proc Natl Acad Sci U S A. 1992 Jan 1;89(1):192–196. doi: 10.1073/pnas.89.1.192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma S. B., Signer E. R. Temporal and spatial regulation of the symbiotic genes of Rhizobium meliloti in planta revealed by transposon Tn5-gusA. Genes Dev. 1990 Mar;4(3):344–356. doi: 10.1101/gad.4.3.344. [DOI] [PubMed] [Google Scholar]
- Spaink H. P., Aarts A., Stacey G., Bloemberg G. V., Lugtenberg B. J., Kennedy E. P. Detection and separation of Rhizobium and Bradyrhizobium Nod metabolites using thin-layer chromatography. Mol Plant Microbe Interact. 1992 Jan-Feb;5(1):72–80. doi: 10.1094/mpmi-5-072. [DOI] [PubMed] [Google Scholar]
- Spaink H. P., Sheeley D. M., van Brussel A. A., Glushka J., York W. S., Tak T., Geiger O., Kennedy E. P., Reinhold V. N., Lugtenberg B. J. A novel highly unsaturated fatty acid moiety of lipo-oligosaccharide signals determines host specificity of Rhizobium. Nature. 1991 Nov 14;354(6349):125–130. doi: 10.1038/354125a0. [DOI] [PubMed] [Google Scholar]
- Spurr A. R. A low-viscosity epoxy resin embedding medium for electron microscopy. J Ultrastruct Res. 1969 Jan;26(1):31–43. doi: 10.1016/s0022-5320(69)90033-1. [DOI] [PubMed] [Google Scholar]
- Surin B. P., Downie J. A. Characterization of the Rhizobium leguminosarum genes nodLMN involved in efficient host-specific nodulation. Mol Microbiol. 1988 Mar;2(2):173–183. doi: 10.1111/j.1365-2958.1988.tb00019.x. [DOI] [PubMed] [Google Scholar]
- Wu H. C., Wu T. C. Isolation and characterization of a glucosamine-requiring mutant of Escherichia coli K-12 defective in glucosamine-6-phosphate synthetase. J Bacteriol. 1971 Feb;105(2):455–466. doi: 10.1128/jb.105.2.455-466.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]