Abstract
Viruses are an integral component of the marine food web, contributing to the disease and mortality of essentially every type of marine life, yet the diversity of viruses in the sea, especially those with RNA genomes, remains very poorly characterized. Isolates of RNA-containing viruses that infect marine plankton are still rare, and the only cultivation-independent surveys of RNA viral diversity reported so far were conducted for temperate coastal waters of British Columbia. Here, we report on our improvements to a previously used protocol to investigate the diversity of marine picorna-like viruses and our results from applying this protocol in subtropical waters. The original protocol was simplified by using direct filtration, rather than tangential flow filtration, to harvest viruses from seawater, and new degenerate primers were designed to amplify a fragment of the RNA-dependent RNA polymerase gene by reverse transcription-PCR from RNA extracted from the filters. Whereas the original protocol was unsuccessful in a preliminary test, the new protocol resulted in amplification of picorna-like virus sequences in every sample of subtropical and temperate coastal seawater assayed. These polymerase sequences formed a diverse, but monophyletic cluster along with other sequences amplified previously from seawater and sequences from isolates infecting marine protists. Phylogenetic analysis suggested that our sequences represent at least five new genera and 24 new species of RNA viruses. These results contribute to our understanding of RNA virus diversity and suggest that picorna-like viruses are a source of mortality for a wide variety of marine protists.
Viruses are an integral component of the marine food web, contributing to the disease and mortality of essentially every type of marine life (11). Although viral genomes may be composed of DNA or RNA and be single stranded (ss) or double stranded (ds), the prevalence of different genome types varies among major groups of organisms that serve as hosts (8). Indirect evidence has suggested that dsDNA viruses are the most abundant form in seawater (18). This is in accordance with the assumption that most free viral particles in seawater (virioplankton) are bacteriophages, a group dominated by representatives having dsDNA genomes (1). Marine algae are also commonly infected with dsDNA viruses in the family Phycodnaviridae, in contrast to land plants, which are almost exclusively infected by viruses having ssRNA genomes (9). Several recent isolations of ssRNA viruses infecting marine protists (4, 14, 15, 17), however, serve as a reminder that numerical abundance is not the best measure of ecological importance and that the diversity of marine virioplankton remains very poorly characterized.
Most efforts to characterize genetic diversity among the virioplankton have focused on DNA viruses. The only published surveys of RNA viral diversity so far have been surveys of temperate coastal waters of British Columbia (5, 7). The first of these studies targeted viruses belonging to the picorna-like virus “superfamily,” which is a group of positive-sense ssRNA viruses that have similar genome features and share conserved regions in several nonstructural genes, including the RNA-dependent RNA polymerase (RdRP) gene (P. Christian, C. M. Fauquet, A. E. Gorbalenya, A. M. G. King, N. Knowles, O. LeGall, and G. Stanway, presented at Microbes in a Changing World, San Francisco, CA, 23 to 28 July 2005). Based on the analysis of conserved RdRP sequences amplified from marine viral assemblages, it was demonstrated (5) that a distantly related group of novel picorna-like viruses are detectable and persistent in the marine environment and that the RdRP gene is a useful molecular marker for the determination of marine RNA virus diversity.
In a subsequent study of the same location, whole-genome shotgun libraries were constructed from the RNA virioplankton of two samples (7). While single-gene surveys can reveal novel variants of known genes, the whole-genome shotgun approach provides a more global assessment of the diversity in a community. Libraries were dominated by RdRP sequences that formed a diverse but monophyletic clade along with homologous sequences from the protistan viruses Heterosigma akashiwo RNA virus (HaRNAV) (10), Rhizosolenia setigera RNA virus (RsRNAV) (12), and Schizochytrium sp. ssRNA virus (SssRNAV) (15). The repeated detection of picorna-like viruses from coastal British Columbia with two independent methods suggests that picorna-like viruses are a consistent component of the RNA virioplankton in that area.
In this study, we wished to determine whether RdRP genes from picorna-like viruses are also detectable in coastal subtropical waters and, if so, how the sequences compare to those from the highly productive temperate waters of British Columbia. Our investigations showed that novel RdRP sequences are readily detectable in Hawaiian waters. Phylogenetic analysis suggested that these gene sequences represent new species and genera of RNA viruses, presumably infecting marine protists.
MATERIALS AND METHODS
Station description.
Water samples were collected with Niskin bottles from March to July 2006 from three environments: Kāneohe Bay, a reef-protected embayment on the windward side of Oahu, Hawaii; Ala Wai Canal, an estuarine urban waterway in Honolulu, HI; and Monterey Bay, a productive cold water embayment located on the central California coast. Samples were collected from three sites in Kāneohe Bay on each of two occasions and analyzed, and a single sample each from the Ala Wai Canal and Monterey Bay was analyzed (Table 1).
TABLE 1.
Station details
| Station | Location | Collection date (mo/day/yr) | Depth (m) | Latitude (°N)a | Longitude (°W)a | Prefilteredb | Sample vol (ml) |
|---|---|---|---|---|---|---|---|
| AWC | Ala Wai Canal | 5/29/2006 | <1 | 21.282383 | 157.826628 | Yes | 250 |
| SB | Kāneohe Bay | 3/2/2006 | <1 | 21.436536 | 157.777361 | No | 50 |
| D | Kāneohe Bay | 3/2/2006 | <1 | 21.423050 | 157.782967 | No | 50 |
| E | Kāneohe Bay | 3/2/2006 | <1 | 21.419067 | 157.780667 | No | 50 |
| AR | Kāneohe Bay | 6/28/2006 | <1 | 21.467639 | 157.836528 | Yes | 250 |
| NR2 | Kāneohe Bay | 6/28/2006 | <1 | 21.515000 | 157.809167 | Yes | 550 |
| SB | Kāneohe Bay | 6/28/2006 | <1 | 21.436536 | 157.777361 | Yes | 350 |
| MBAY | Monterey Bay | 7/22/2006 | 16 | 36.935133 | 121.918600 | Yes | 200 |
| MBAY | Monterey Bay | 7/22/2006 | 16 | 36.935133 | 121.918600 | No | 125 |
Latitude and longitude are indicated using decimal coordinates.
Sterivex (Millipore) 0.22-μm-pore-size in-line filters were used where indicated.
Collection.
Water samples collected from Monterey Bay, from the Ala Wai Canal, and from stations NR2, AR, and SB in Kāneohe Bay in July 2006 were pressure filtered (7 mm Hg), using a peristaltic pump, through a 0.22-μm-pore-size polyether sulfone membrane filter cartridge (Sterivex; Millipore, Billerica, MA), followed by a 0.02-μm aluminum oxide filter (Anotop; Whatman, Middlesex, United Kingdom). In the latter case, filtration was continued until the filtration rate decreased dramatically or dropped to zero (200 to 550 ml). Whole seawater samples collected from stations SB, D, and E in Kāneohe Bay in March 2006 were filtered directly onto 0.02-μm aluminum oxide filters in the same manner but were limited to 50 ml. After processing, the filter inlets and outlets were sealed, and the filters were stored at −80°C until extraction.
Extraction.
Total nucleic acids were extracted from the aluminum oxide filters using a Masterpure complete DNA and RNA purification kit (Epicenter, Madison, WI) with a protocol slightly modified from the manufacturer's instructions. Briefly, 400 μl of 2× T+C lysis buffer containing 50 μg/μl proteinase K was added to a 3-ml syringe. The syringe was locked to the filter inlet, and the lysis buffer was gently pushed into the filter until the buffer just reached the outlet. A flame-sealed, sterile, 1,000-μl pipette tip was firmly inserted into the filter outlet, and the entire assembly was incubated for 10 min at 65°C in air. Afterward, the filter tip was removed, and the extracted material was gently evacuated through the outlet into a sterile microcentrifuge tube by applying pressure with the syringe. Extracts were immediately placed on ice. Salt-induced precipitation of protein-detergent complexes followed by alcohol precipitation and washing of total nucleic acids was carried out according to the manufacturer's instructions.
DNase treatment.
Precipitated total nucleic acids were resuspended in 10 μl Tris-EDTA buffer and then incubated with 1 U DNase I (Invitrogen, Carlsbad, CA) and 1× DNase I buffer for 15 min at room temperature to remove DNA from the sample. The reaction was terminated by adding 2.5 mM (final concentration) EDTA and incubating the preparation for 10 min at 65°C.
Reverse transcription (RT)-PCR.
(i) Primer design. Conserved regions of the putative RdRP sequences from the marine picorna-like viruses (5) were aligned using CLUSTAL X v1.83 with the Gonnet series protein matrix (16). The alignment was used to design four degenerate primer sets with various specificities. Three of the primer sets were designed to target subclades 1 to 3 (Mpl.sc1, Mpl.sc2, and Mpl.sc3) within the larger cluster but overlapped in their target ranges. A fourth primer set (Mpl.cdh) was designed based on the consensus degenerate hybrid oligonucleotide (CODEHOP) strategy (13) with the aim of capturing broader diversity across, and possibly outside, the cluster as it was defined at the time.
(ii) cDNA synthesis.
cDNA was synthesized using reverse transcriptase (Superscript III; Invitrogen, Carlsbad, CA) primed with random hexamers, Mpl.sc1R, Mpl.sc2R, Mpl.sc3R, or Mpl.cdhR. The reaction mixtures (total volume, 13 μl) contained 8 μl of extracted DNase-treated RNA template, 0.2 mM of each deoxynucleoside triphosphate, and either 100 ng (N6) or 5 pmol (all other primers) of primer. Samples were denatured at 65°C and then cooled on ice, at which point the reaction mixtures were supplemented with dithiothreitol (final concentration, 0.5 mM), 40 U RNase OUT (Invitrogen), and 200 U Superscript III (Invitrogen) to obtain a final reaction mixture volume of 20 μl. The reaction mixtures were mixed and brought to 25°C for 5 min, and this was followed by incubation at 50°C for 55 min (N6-primed reactions) or at the primer-specific annealing temperature (Table 2) for 45 min. All samples were incubated at 70°C for 15 min as a final termination step.
TABLE 2.
RT and PCR primers
| Primer | Sequence (5′-3′) | Annealing temp (°C) | Approx product size (bp) | Genome(s) based ona |
|---|---|---|---|---|
| Mpl.sc1F | TIGCIGGWGAYTWYARM | 50 | 500 | JP-A, JP-B, RsRNAV |
| Mpl.sc1R | YTCCTTWTCRGSCATKGTA | |||
| Mpl.sc2F | ITWGCIGGIGATTWCA | 43 | 500 | HaRNAV, JP-A, RsRNAV |
| Mpl.sc2R | CKYTTCARRAAWTCAGCATC | |||
| Mpl.sc3F | TIATIGMKGGIGAYTA | 49 | 500 | HaRNAV, JP-A, SssRNAV |
| Mpl.sc3R | TTMARGAAIKMAGCATCTT | |||
| Mpl.cdhF | GMIGGTGAYTAYAGCGCTTWYGAY | 44 | 500 | JP-A, JP-B, RsRNAV, HaRNAV, SssRNAV |
| Mpl.cdhR | ATACCCAATGCCTYTTIARRAA |
The primers were designed from an alignment of the genomes listed.
(iii) PCR.
PCR was performed with the Mpl.sc1, Mpl.sc2, Mplsc3, and Mpl.cdh primer sets listed in Table 2. Each reaction mixture (final volume, 50 μl) consisted of 1× Platinum Taq buffer, 3 mM MgCl2, 0.2 mM of each deoxynucleoside triphosphate, 1 μM of each primer, and 1 U Platinum Taq DNA polymerase (Invitrogen). The following thermocycler conditions were used: 94°C for 75 s, followed by 40 cycles of denaturation at 94°C for 45 s, annealing at the primer-specific temperature (Table 2) for 45 s, and extension at 72°C for 1 min and then a final extension step of 9 min at 72°C. PCR products were separated on a 1.5% agarose gel, and DNA of the appropriate size (approximately 500 bp) was excised and purified using a Minelute gel extraction kit (QIAGEN, Valencia, CA) according to the manufacturer's instructions.
(iv) Cloning and sequencing.
Purified PCR products were cloned directly into the TOPO-TA vector (Invitrogen) and transformed into TOP10 DH5α competent cells (Invitrogen) according to instructions provided with the vector and cells. Clones were screened for inserts by PCR amplification with the universal M13 forward (−20; 5′-GTAAAACGACGGCCAGT-3′) and M13 reverse (−27; 5′-CAGGAAACAGCTATGAC-3′) primers. Amplified products of the correct size were sequenced by the ASGPB sequencing service at the University of Hawaii at Mānoa with the M13 reverse primer.
(v) Phylogenetic analyses.
Sequences that were more than 98% identical at the nucleotide level were considered a single phylotype. Phylotypes were compared to sequences in the NCBI database with tBLASTx (3). Details of the viruses and environmental sequences used in phylogenetic analyses are shown in Table S1 in the supplemental material. Translated sequences of viruses were aligned using CLUSTAL X v1.83 with the Gonnet series protein matrix (16). Alignments were transformed into likelihood distances by using Mr Bayes v3.1.2 (2) and 1,000,000 generations.
Nucleotide sequence accession numbers.
Sequences have been deposited in the GenBank database, and the accession numbers are listed in Table S1 in the supplemental material.
RESULTS
Viral RdRP gene sequences were successfully amplified using as the template RNA extracted from marine plankton collected by direct filtration onto 25-mm, 0.02-μm-pore-size aluminum oxide filters. In one case (Monterey Bay sample), amplification was not successful when a whole-plankton extract was used but was successful with an extract from a parallel sample that had been prefiltered through a 0.22-μm-pore-size filter prior to collection on a 0.02-μm filter. Amplification occurred for several distinct aquatic environments, including an estuarine urban canal and both subtropical and temperate bays. Furthermore, amplification occurred with samples from the same site obtained in different seasons and at different depths. All sequenced environmental PCR products translated into continuous amino acid sequences that contained the motif GDD, which is present in almost all positive-sense ssRNA virus polymerases. Using tBLASTx (3), the top-scoring matches in GenBank to the environmental sequences from this study were, without exception, putative RdRP sequences from picorna-like viruses.
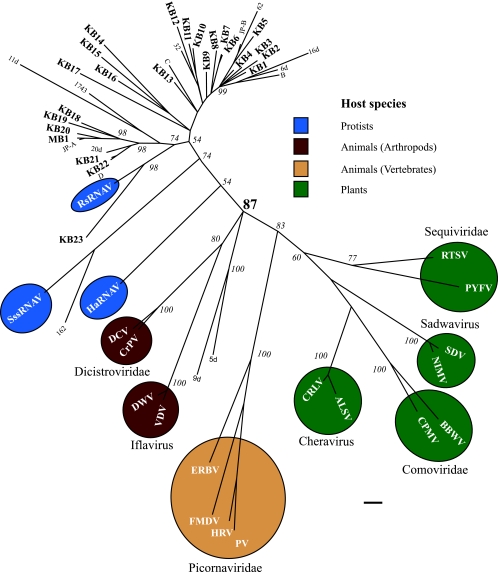
A Bayesian phylogenetic analysis based on an amino acid sequence alignment resulted in single large clade with a Bayesian support value of 87 that contained all of the environmental RdRP sequences (phylotypes) from this study, RdRP sequences obtained from seawater samples collected in coastal British Columbia, Canada, and the RdRP sequences of three RNA virus isolates that infect marine protists (Fig. 1). Established families of picorna-like viruses that infect plants and animals were all resolved into monophyletic clusters with Bayesian support values of 77 to 100 (Fig. 1). The levels of amino acid identity of phylotypes to their nearest neighbors in the tree ranged from 38 to 98%, and the two phylotypes recovered in this study that were most divergent from one another (KB15 and KB23) shared only 24% amino acid identity.
FIG. 1.
Phylogenetic tree of picorna-like virus RdRP sequences: Bayesian maximum likelihood tree for RdRP amino acid sequences in the picorna-like virus superfamily. Regions of the tree are colored coded to indicate host type where known. The designations for sequences retrieved in this study are in bold type and begin with “KB” (for Kāneohe Bay sequences) or “MB” (for Monterey Bay sequences). Sequences designated B to D, sequences designated with numbers only, and the sequences designated JP-A and JP-B are environmental sequences from previously published investigations (5, 7). Amino acid alignments were transformed into likelihood distances with Mr Bayes v3.1.1 (2) using 1,000,000 generations. Bayesian clade credibility values are shown for relevant nodes. The Bayesian scale bar indicates a distance of 0.1. RTSV, rice tungro spherical virus; PYFV, parsnip yellow fleck virus; SDV, satsuma dwarf virus; NIMV, naval orange infectious mottling virus; BBWV, broad bean wilt virus; CPMV, cowpea mosaic virus; ALSV, apple latent spherical virus; CRLV, cherry rasp leaf virus; ERBV, equine rhinitis B virus; FMDV, foot-and-mouth disease virus; HRV, human rhinovirus; PV, poliovirus; DWV, deformed wing virus; VDV, Varroa destructor virus; DCV, Drosophila C virus; CrPV, cricket paralysis virus.
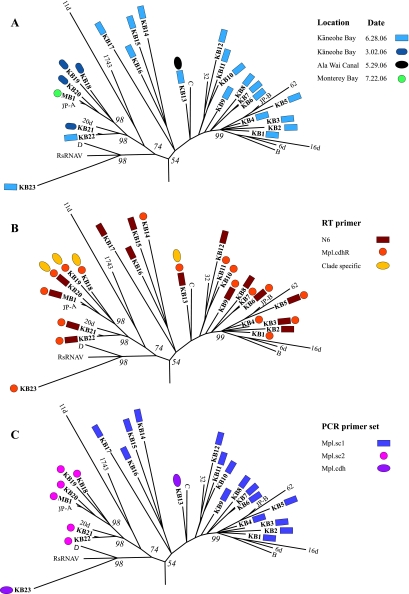
The richness of phylotypes detected in a single sample varied, ranging from 1 to 19 phylotypes (Fig. 2A), and 24 phylotypes were detected overall. Identical amino acid sequences were recovered from two different samples in only one instance; phylotype KB13 was recovered from Kāneohe Bay, a site characterized by high salinity and low nutrients, and from the Ala Wai Canal, a eutrophic estuarine drainage canal in the city of Honolulu (Fig. 2A). No identical phylotypes were retrieved from samples obtained in Kāneohe Bay on two different occasions. The two most similar sequences, KB21 and KB22 from these samples, shared 86% identity. Phylotype MB1, amplified from a sample collected in Monterey Bay in California in 2006, shared 97% amino acid identity with the homologous region of an RdRP recovered from coastal British Columbia in 2000 (7).
FIG. 2.
Relationship between the distribution of phylotypes recovered and the sample source or the primers used for RT or PCR amplification. The Mpl clade, clipped from the tree constructed in Fig. 1, is displayed with each phylotype recovered in this study coded by shape and color to indicate the location and date of each sample (A), the RT primer(s) used that produced the phylotype (B), and the PCR primer set(s) that yielded the phylotype (C).
Although an initial attempt to amplify picorna-like virus sequences from our samples using the methods described by Culley et al. (5) was unsuccessful, use of the new RT primers designed as part of this study resulted in subsequent amplification with at least one of the newly designed PCR primer sets. Together, the clade-specific RT primers captured only 4 of the 24 phylotypes (Fig. 2B). Each of these sequences was also recovered when RT reactions were primed with the less specific Mpl.cdhR primer or the nonspecific N6 primer. Of the 24 clades recovered, 7 were unique to cDNA reactions primed with Mpl.cdhR and 6 were unique to reactions primed with N6, while 9 clades were amplified with both primers (Fig. 2B).
PCR amplification with the clade-specific primers Mpl.sc1 and Mpl.sc2 resulted in two distinct clusters of sequences, which together accounted for 22 of the 24 phylotypes detected (Fig. 2C). Mpl.sc3 failed to amplify a product in all of the samples tested. The remaining two phylotypes were detected only by the Mpl.cdh primers (Fig. 2C).
The relative abundances of different phylotypes recovered were compared for two clone libraries produced from the same sample (Kāneohe Bay station SB), using the same PCR primers (Mpl.sc1) but different methods of cDNA synthesis (primer N6 or Mpl.cdhR). Phylotype KB3 dominated both libraries, accounting for 45% (10/28) of the sequences in the N6-primed library and 36% (8/22) of the sequences in the Mpl.cdhR-primed sample. Of the 16 phylotypes detected, 4 were present in both libraries, while 6 were unique to each library. Of the 12 unique phylotypes, 10 were represented by one sequence.
DISCUSSION
This study more than doubled the number of phylotypes in a cluster of diverse sequences referred to as the “marine picorna-like” (Mpl) virus clade (7). Our results provide further support for the idea that this group is monophyletic and distantly related to the previously established taxa of picorna-like viruses that infect higher plants and animals. The few viral isolates whose RdRP sequences cluster within this new major clade are viruses of marine protists, but whether it is appropriate that this clade be referred to as “marine” remains to be seen. All of the sequences in this clade to date have been derived from marine sources, but freshwater samples have not been screened with the same methods yet. Furthermore, RdRP sequences of International Committee on the Taxonomy of Viruses (ICTV)-recognized picorna-like viruses cluster in accordance with phylogenetic relationships among their hosts rather than as a function of environment per se. We hypothesize, therefore, that this new and expanding clade will ultimately be better described as a protistan picorna-like virus clade, but additional isolates and data are needed to confirm this proposition. For now, we defer to precedent (7) and use the designation “marine picorna-like” or Mpl clade.
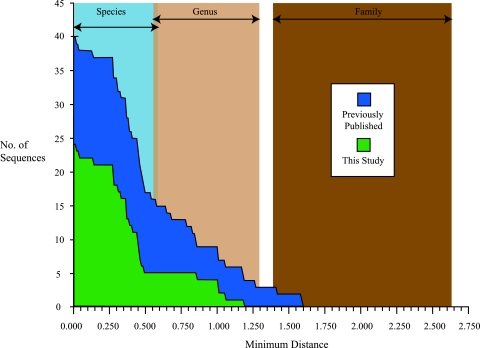
There is considerable diversity within the Mpl clade, suggesting that a wide range of marine protists are infected with picorna-like viruses. If we assume that the relationship between RdRP gene sequence divergence and phylogenetic affiliation is roughly consistent across the picorna-like viruses, then the distances between known phylogenetic groups can be used to infer the level of taxonomic diversity represented among the Mpl viruses. Such an analysis suggests that, at present, the Mpl clade consists of at least three families composed of 16 genera and 40 species (Fig. 3). This level of diversity is comparable to the existing taxa of picorna-like viruses that are officially recognized at present (Christian et al., presented at Microbes in a Changing World, 23 to 28 July 2005).
FIG. 3.
Assessment of phylogenetic divergence within the marine picorna-like clade based on Bayesian distances: cumulative frequency distribution of the minimum Bayesian distances between each unique, partial RdRP sequence in the Mpl clade and its nearest neighbor. For each distance step of 0.01, the number of sequences in the Mpl clade at that or a greater distance from its nearest neighbor is plotted. The distribution is divided to show the contribution of the new sequences from this study to the total number of known sequences within the Mpl clade. Labeled regions indicate the ranges of Bayesian distance that constitute family, genus, and species level divergence within currently established taxa of picorna-like viruses recognized by the ICTV. Specifically, the indicated ranges are based on the calculated Bayesian distances among all available homologous partial RdRP sequences for members of three families of picorna-like viruses (Comoviridae, Picornaviridae, and Sequiviridae). These three families were selected for analysis because each of them also includes ICTV-recognized genera and species. The upper value for each range is the shortest calculated distance between any two RdRP sequences derived from viruses that are considered to be members of different taxa. The lower value for each range is the greatest distance calculated between any two RdRP sequences derived from viruses that are still considered to be members of the same taxon. Multiple-sequence alignment and calculation of Bayesian distances were carried out as described in Materials and Methods.
Of the three putative Mpl families, one is the previously described family Marnaviridae, which consists, so far, of only HaRNAV. Our analysis, therefore, supports the recent recognition of this isolate as the representative of a new family in a report by the ICTV (6). The second family is represented by a thraustochytrid virus isolate, SssRNAV, along with a single related environmental sequence. The third family consists of many diverse sequences obtained from seawater by cultivation-independent methods along with the sole cultivated representative, RsRNAV, which infects a diatom. The sequences retrieved in this study represent addition of up to 24 species and five genera of picorna-like viruses to the latter family.
The detection of an RdRP sequence from Monterey Bay in California that is very similar to a sequence recovered 6 years earlier from English Bay in British Columbia ca. 1,500 km to the north (5) suggests the presence of at least one picorna-like virus species that is persistent and relatively widespread in temperate coastal waters. In contrast, the lack of common sequences in samples from subtropical coastal waters of Oahu and temperate coastal waters of the North Pacific most likely reflects differences in the plankton community composition between these two habitats. The sample coverage and sequence coverage are still too low, however, to draw firm conclusions about the biogeography of picorna-like viruses.
The failure of the previously described RT-PCR protocol described by Culley et al. (5) to amplify RdRP sequences from a coastal Oahu sample suggested initially that picorna-like viruses were either absent or substantially different from those recovered from coastal British Columbia. Since the primers used by Culley et al. (5) were designed from the very limited number of sequences available at the time, we opted to design new sets of RT and PCR primers with various specificities, incorporating new information about marine picorna-like RdRP sequences, rather than to further test the original primer sets. The results obtained from use of these new primer sets in various combinations provide some insight into the most effective strategies for detecting RdRP genes from the environment.
The use of broad-specificity (Mpl.cdh) or nonspecific (N6) primers for the RT step resulted in recovery of the greatest sequence diversity. Some unique sequences were obtained with each of these primers, but they retrieved sequences over similarly broad phylogenetic ranges with substantial overlap. The data are insufficient to conclude whether the minor differences between them are significant. The Mpl.cdh primer, designed to prime RT of diverse RdRP sequences, might be useful for boosting the signal-to-noise ratio in cases where there is a very high background of nontarget RNA. However, given that the completely nonspecific N6 primer was effective even with samples that had not been prefiltered, this may be the best primer to use for the RT step, particularly since the range of RdRP diversity in the environment is still so poorly known.
Since it was not practical to design a single degenerate PCR primer set that could encompass the diversity within the entire Mpl clade, we used two strategies to maximize the range of sequences retrieved. The use of degenerate, subclade-targeted PCR primer sets Mpl.sc1 and Mpl.sc2 resulted in selective amplification of sequences that clustered in different portions of the Mpl clade, as was expected based on their design. The failure of Mpl.sc3 to amplify any targets suggests that the HaRNAV- and SssRNAV-like sequences were not common in our samples.
The consensus degenerate hybrid primer set, Mpl.cdh, retrieved only two distinct phylotypes, neither of which was recovered with the subclade primers. The hybrid primers are capable, in principle, of retrieving the sequences of novel, highly divergent members of a protein family because the 3′ end is fully degenerate for short, conserved amino acid motifs. However, because the 5′ portion of the primers is not degenerate, these primers are also likely to exhibit significant amplification bias in the presence of mixed targets. These properties may explain why the Mpl.cdh PCR primers recovered rather divergent sequences but very few sequences overall. The apparent lack of bias when the Mpl.cdhR primer was used for RT might be attributable to the fundamental differences between the kinetics of an RT reaction and those of a PCR. Since each of the four PCR primer sets appeared to target a different subset of the Mpl clade, we recommend screening samples with all four of the primer sets in order to increase the diversity of Mpl RdRP sequences recovered from environmental samples.
A major processing bottleneck and possible source of bias in previous studies of viral RdRP diversity has been the use of prefiltration followed by tangential flow ultrafiltration. Here we demonstrated that it is possible to recover RdRP sequences from virioplankton collected by direct filtration of relatively small volumes of seawater onto 0.02-μm-pore-size filters. In one case (Monterey Bay sample), amplification was successful only when the sample was prefiltered, perhaps due to the presence of PCR inhibitors in the whole-water extract. However, successful amplifications of RdRP were achieved for a number of other samples with no prefiltration. This suggests that direct filtration of whole seawater is a reasonable sample collection strategy that avoids the pitfalls associated with prefiltration, although in some cases additional steps to remove PCR inhibitors may be required.
Direct filtration involves less sample manipulation and is faster than tangential flow ultrafiltration, which should help to minimize some sources of bias, such as selective losses of viruses due to trapping on prefilters or decay of labile viruses during prolonged processing of concentrates. This procedure also avoids the economic and operational drawbacks of tangential flow filtration, namely, the expense of the equipment and the use of filters that must be cleaned between samples. The rapidity and simplicity of direct filtration should allow investigations of marine RNA viral diversity at greater temporal and spatial resolution than is possible when tangential flow ultrafiltration is employed.
In summary, we successfully applied a simplified virus sampling protocol along with a suite of new primers to recover novel picornavirus-like RdRP gene sequences from samples of subtropical and temperate waters. The results extend the known geographic range of picorna-like viruses in seawater and contribute substantially to the known RdRP sequence diversity. Phylogenetic analysis suggested that the recently described and very diverse clade of “marine picorna-like” viruses most likely represents a wealth of protistan viruses which have yet to be isolated and described but which are likely to have a significant influence on the composition and dynamics of the marine eukaryotic plankton.
Supplementary Material
Acknowledgments
We thank Olivia Nigro, Sara Yeo, and Misty Miller for their help with sample collection in Kāneohe Bay and the participants in the ONR-funded program Layered Organization in the Coastal Ocean and Jim Christian, Captain of the R/V Shana Rae, for their assistance with sampling in Monterey Bay.
This work was supported by grants OCE 06-0026 and EF 04-24599 from the National Science Foundation and by grant UAF06-0026 from the NOAA West Coast & Polar Regions Undersea Research Center.
Footnotes
Published ahead of print on 20 July 2007.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Ackermann, H. W. 2007. 5500 Phages examined in the electron microscope. Arch. Virol. 152:227-243. [DOI] [PubMed] [Google Scholar]
- 2.Altekar, G., S. Dwarkadas, J. P. Huelsenbeck, and F. Ronquist. 2004. Parallel metropolis coupled Markov chain Monte Carlo for Bayesian phylogenetic inference. Bioinformatics 20:407-415. [DOI] [PubMed] [Google Scholar]
- 3.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. H. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brussaard, C. P. D., A. A. M. Noordeloos, R. A. Sandaa, M. Heldal, and G. Bratbak. 2004. Discovery of a dsRNA virus infecting the marine photosynthetic protist Micromonas pusilla. Virology 319:280-291. [DOI] [PubMed] [Google Scholar]
- 5.Culley, A. I., A. S. Lang, and C. A. Suttle. 2003. High diversity of unknown picorna-like viruses in the sea. Nature 424:1054-1057. [DOI] [PubMed] [Google Scholar]
- 6.Culley, A. I., A. S. Lang, and C. A. Suttle. 2004. Marnaviridae, p. 789-792. In C. M. Fauquet, M. A. Mayo, J. Maniloff, U. Desselberger, and L. A. Ball (ed.), Virus taxonomy: eighth report of the International Committee on Taxonomy of Viruses. Academic Press, Oxford, United Kingdom.
- 7.Culley, A. I., A. S. Lang, and C. A. Suttle. 2006. Metagenomic analysis of coastal RNA virus communities. Science 312:1795-1798. [DOI] [PubMed] [Google Scholar]
- 8.Dimmock, N. J., and S. B. Primrose. 1994. Introduction to modern virology, 4th ed. Blackwell Science, Cambridge, MA.
- 9.Dunigan, D. D., L. A. Fitzgerald, and J. L. Van Etten. 2006. Phycodnaviruses: A peek at genetic diversity. Virus Res. 117:119-132. [DOI] [PubMed] [Google Scholar]
- 10.Lang, A. S., A. I. Culley, and C. A. Suttle. 2004. Genome sequence and characterization of a virus (HaRNAV) related to picorna-like viruses that infects the marine toxic bloom-forming alga Heterosigma akashiwo. Virology 320:206-217. [DOI] [PubMed] [Google Scholar]
- 11.Munn, C. B. 2006. Viruses as pathogens of marine organisms—from bacteria to whales. J. Mar. Biol. Assoc. U. K. 86:453-467. [Google Scholar]
- 12.Nagasaki, K., Y. Tomaru, N. Katanozaka, Y. Shirai, K. Nishida, I. Shigeru, and M. Yamaguchi. 2004. Isolation and characterization of a novel single-stranded RNA virus infecting the bloom-forming diatom Rhizosolenia setigera. Appl. Environ. Microbiol. 70:704-711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rose, T. M., E. R. Schultz, J. G. Henikoff, S. Pietrokovski, C. M. McCallum, and S. Henikoff. 1998. Consensus-degenerate hybrid oligonucleotide primers for amplification of distantly related sequences. Nucleic Acids Res. 26:1628-1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tai, V., J. E. Lawrence, A. S. Lang, A. M. Chan, A. I. Culley, and C. A. Suttle. 2003. Characterization of HaRNAV, a single-stranded RNA virus causing lysis of Heterosigma akashiwo (Raphidophyceae). J. Phycol. 39:343-352. [Google Scholar]
- 15.Takao, Y., K. Nagasaki, K. Mise, T. Okuno, and D. Honda. 2005. Isolation and characterization of a novel single-stranded RNA virus infectious to a marine fungoid protist, Schizochytrium sp. (Thraustochytriaceae, Labyrinthulea). Appl. Environ. Microbiol. 71:4516-4522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The CLUSTAL_X Windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25:4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tomaru, Y., N. Katanozaka, K. Nishida, Y. Shirai, K. Tarutani, M. Yamaguchi, and K. Nagasaki. 2004. Isolation and characterization of two distinct types of HcRNAV, a single-stranded RNA virus infecting the bivalve-killing microalga Heterocapsa circularisquama. Aquat. Microb. Ecol. 34:207-218. [Google Scholar]
- 18.Weinbauer, M. G. 2004. Ecology of prokaryotic viruses. FEMS Microbiol. Rev. 28:127-181. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.