Abstract
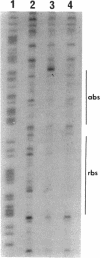
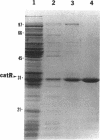
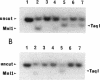
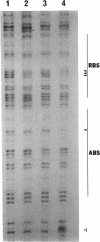
In Pseudomonas putida, the catBC operon encodes enzymes involved in benzoate degradation. Previous studies have determined that these enzymes are induced when P. putida is grown in the presence of benzoate. Induction of the enzymes of the catBC operon requires an intermediate of benzoate degradation, cis,cis-muconate, and a regulatory protein, CatR. It has been determined that CatR binds to a 27-bp region of the catBC promoter in the presence or absence of inducer. We have called this the repression binding site. In this study, we used a gel shift assay to demonstrate that the inducer, cis,cis-muconate, increases the affinity of CatR for the catBC promoter region by 20-fold. Furthermore, in the absence of cis,cis-muconate, CatR forms two complexes in the gel shift assay. The inducer cis,cis-muconate confers specificity primarily for the formation of complex 2. DNase I footprinting showed that an additional 27 bp of the catBC promoter region is protected by CatR in the presence of cis,cis-muconate. We have named this second binding site the activation binding site. Methylation interference footprinting determined that in the presence or absence of inducer, five G nucleotides of the catBC promoter region were necessary for CatR interaction with the repression binding site, while a single G residue was important for CatR interaction with the activation binding site in the presence of cis,cis-muconate. Using polymerase chain reaction-generated constructs, we found that the binding of CatR to the repression binding site is independent of the activation binding site. However, binding of CatR to the activation binding site required an intact repression binding site.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Aldrich T. L., Chakrabarty A. M. Transcriptional regulation, nucleotide sequence, and localization of the promoter of the catBC operon in Pseudomonas putida. J Bacteriol. 1988 Mar;170(3):1297–1304. doi: 10.1128/jb.170.3.1297-1304.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aldrich T. L., Frantz B., Gill J. F., Kilbane J. J., Chakrabarty A. M. Cloning and complete nucleotide sequence determination of the catB gene encoding cis,cis-muconate lactonizing enzyme. Gene. 1987;52(2-3):185–195. doi: 10.1016/0378-1119(87)90045-x. [DOI] [PubMed] [Google Scholar]
- Aldrich T. L., Rothmel R. K., Chakrabarty A. M. Identification of nucleotides critical for activity of the Pseudomonas putida catBC promoter. Mol Gen Genet. 1989 Aug;218(2):266–271. doi: 10.1007/BF00331277. [DOI] [PubMed] [Google Scholar]
- Bradford M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976 May 7;72:248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- Chang M., Crawford I. P. In vitro determination of the effect of indoleglycerol phosphate on the interaction of purified TrpI protein with its DNA-binding sites. J Bacteriol. 1991 Mar;173(5):1590–1597. doi: 10.1128/jb.173.5.1590-1597.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang M., Crawford I. P. The roles of indoleglycerol phosphate and the TrpI protein in the expression of trpBA from Pseudomonas aeruginosa. Nucleic Acids Res. 1990 Feb 25;18(4):979–988. doi: 10.1093/nar/18.4.979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook A. M., Grossenbacher H., Hütter R. Isolation and cultivation of microbes with biodegradative potential. Experientia. 1983 Nov 15;39(11):1191–1198. doi: 10.1007/BF01990356. [DOI] [PubMed] [Google Scholar]
- Fried M., Crothers D. M. Equilibria and kinetics of lac repressor-operator interactions by polyacrylamide gel electrophoresis. Nucleic Acids Res. 1981 Dec 11;9(23):6505–6525. doi: 10.1093/nar/9.23.6505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galas D. J., Schmitz A. DNAse footprinting: a simple method for the detection of protein-DNA binding specificity. Nucleic Acids Res. 1978 Sep;5(9):3157–3170. doi: 10.1093/nar/5.9.3157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao J., Gussin G. N. Mutations in TrpI binding site II that differentially affect activation of the trpBA promoter of Pseudomonas aeruginosa. EMBO J. 1991 Dec;10(13):4137–4144. doi: 10.1002/j.1460-2075.1991.tb04991.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henikoff S., Haughn G. W., Calvo J. M., Wallace J. C. A large family of bacterial activator proteins. Proc Natl Acad Sci U S A. 1988 Sep;85(18):6602–6606. doi: 10.1073/pnas.85.18.6602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hryniewicz M. M., Kredich N. M. The cysP promoter of Salmonella typhimurium: characterization of two binding sites for CysB protein, studies of in vivo transcription initiation, and demonstration of the anti-inducer effects of thiosulfate. J Bacteriol. 1991 Sep;173(18):5876–5886. doi: 10.1128/jb.173.18.5876-5886.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J. Z., Schell M. A. In vivo interactions of the NahR transcriptional activator with its target sequences. Inducer-mediated changes resulting in transcription activation. J Biol Chem. 1991 Jun 15;266(17):10830–10838. [PubMed] [Google Scholar]
- Kato J., Chakrabarty A. M. Purification of the regulatory protein AlgR1 and its binding in the far upstream region of the algD promoter in Pseudomonas aeruginosa. Proc Natl Acad Sci U S A. 1991 Mar 1;88(5):1760–1764. doi: 10.1073/pnas.88.5.1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lindquist S., Lindberg F., Normark S. Binding of the Citrobacter freundii AmpR regulator to a single DNA site provides both autoregulation and activation of the inducible ampC beta-lactamase gene. J Bacteriol. 1989 Jul;171(7):3746–3753. doi: 10.1128/jb.171.7.3746-3753.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meagher R. B., McCorkle G. M., Ornston M. K., Ornston L. N. Inducible uptake system for -carboxy-cis, cis-muconate in a permeability mutant of Pseudomonas putida. J Bacteriol. 1972 Aug;111(2):465–473. doi: 10.1128/jb.111.2.465-473.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messing J., Crea R., Seeburg P. H. A system for shotgun DNA sequencing. Nucleic Acids Res. 1981 Jan 24;9(2):309–321. doi: 10.1093/nar/9.2.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ORNSTON L. N., STANIER R. Y. MECHANISM OF BETA-KETOADIPATE FORMATION BY BACTERIA. Nature. 1964 Dec 26;204:1279–1283. doi: 10.1038/2041279a0. [DOI] [PubMed] [Google Scholar]
- Rothmel R. K., Aldrich T. L., Houghton J. E., Coco W. M., Ornston L. N., Chakrabarty A. M. Nucleotide sequencing and characterization of Pseudomonas putida catR: a positive regulator of the catBC operon is a member of the LysR family. J Bacteriol. 1990 Feb;172(2):922–931. doi: 10.1128/jb.172.2.922-931.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothmel R. K., Shinabarger D. L., Parsek M. R., Aldrich T. L., Chakrabarty A. M. Functional analysis of the Pseudomonas putida regulatory protein CatR: transcriptional studies and determination of the CatR DNA-binding site by hydroxyl-radical footprinting. J Bacteriol. 1991 Aug;173(15):4717–4724. doi: 10.1128/jb.173.15.4717-4724.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schell M. A., Poser E. F. Demonstration, characterization, and mutational analysis of NahR protein binding to nah and sal promoters. J Bacteriol. 1989 Feb;171(2):837–846. doi: 10.1128/jb.171.2.837-846.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siebenlist U., Gilbert W. Contacts between Escherichia coli RNA polymerase and an early promoter of phage T7. Proc Natl Acad Sci U S A. 1980 Jan;77(1):122–126. doi: 10.1073/pnas.77.1.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storz G., Tartaglia L. A., Ames B. N. Transcriptional regulator of oxidative stress-inducible genes: direct activation by oxidation. Science. 1990 Apr 13;248(4952):189–194. doi: 10.1126/science.2183352. [DOI] [PubMed] [Google Scholar]
- Vieira J., Messing J. Production of single-stranded plasmid DNA. Methods Enzymol. 1987;153:3–11. doi: 10.1016/0076-6879(87)53044-0. [DOI] [PubMed] [Google Scholar]
- Wek R. C., Hatfield G. W. Transcriptional activation at adjacent operators in the divergent-overlapping ilvY and ilvC promoters of Escherichia coli. J Mol Biol. 1988 Oct 5;203(3):643–663. doi: 10.1016/0022-2836(88)90199-4. [DOI] [PubMed] [Google Scholar]
- Werel W., Schickor P., Heumann H. Flexibility of the DNA enhances promoter affinity of Escherichia coli RNA polymerase. EMBO J. 1991 Sep;10(9):2589–2594. doi: 10.1002/j.1460-2075.1991.tb07800.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheelis M. L., Ornston L. N. Genetic control of enzyme induction in the -ketoadipate pathway of Pseudomonas putida: deletion mapping of cat mutations. J Bacteriol. 1972 Feb;109(2):790–795. doi: 10.1128/jb.109.2.790-795.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C. H., Ornston M. K., Ornston L. N. Genetic control of enzyme induction in the -ketoadipate pathway of Pseudomonas putida: two-point crosses with a regulatory mutant strain. J Bacteriol. 1972 Feb;109(2):796–802. doi: 10.1128/jb.109.2.796-802.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C. C., Nash H. A. The interaction of E. coli IHF protein with its specific binding sites. Cell. 1989 Jun 2;57(5):869–880. doi: 10.1016/0092-8674(89)90801-5. [DOI] [PubMed] [Google Scholar]