Abstract
The hyperthermophilic archaeon Thermococcus kodakaraensis harbors a type III ribulose 1,5-bisphosphate carboxylase/oxygenase (RbcTk). It has previously been shown that RbcTk is capable of supporting photoautotrophic and photoheterotrophic growth in a mesophilic host cell, Rhodopseudomonas palustris Δ3, whose three native Rubisco genes had been disrupted. Here, we have examined the enzymatic properties of RbcTk at 25°C and have constructed mutant proteins in order to enhance its performance in mesophilic host cells. Initial sites for mutagenesis were selected by focusing on sequence differences in the loop 6 and α-helix 6 regions among RbcTk and the enzymes from spinach (mutant proteins SP1 to SP7), Galdieria partita (GP1 and GP2), and Rhodospirillum rubrum (RR1). Loop 6 of RbcTk is one residue longer than those found in the spinach and G. partita enzymes, and replacing RbcTk loop 6 with these regions led to dramatic decreases in activity. Six mutant enzymes retaining significant levels of Rubisco activity were selected, and their genes were introduced into R. palustris Δ3. Cells harboring mutant protein SP6 displayed a 31% increase in the specific growth rate under photoheterotrophic conditions compared to cells harboring wild-type RbcTk. SP6 corresponds to a complete substitution of the original α-helix 6 of RbcTk with that of the spinach enzyme. Compared to wild-type RbcTk, the purified SP6 mutant protein exhibited a 30% increase in turnover number (kcat) of the carboxylase activity and a 17% increase in the kcat/Km value. Based on these results, seven further mutant proteins were designed and examined. The results confirmed the importance of the length of loop 6 in RbcTk and also led to the identification of specific residue changes that resulted in an increase in the turnover number of RbcTk at ambient temperatures.
Ribulose 1,5-bisphosphate carboxylase/oxygenase (Rubisco; EC 4.1.1.39) is the key enzyme of the Calvin-Benson-Bassham (CBB) pathway. As all plants, algae, and cyanobacteria and many other autotrophic bacteria utilize the CBB pathway, Rubisco can be considered the main gateway for organic carbon production from CO2 on our planet. The carboxylase activity of Rubisco produces two molecules of 3-phosphoglycerate from ribulose 1,5-bisphosphate (RuBP), CO2, and H2O. However, the enzyme also catalyzes a competing oxygenase reaction, converting O2 and RuBP to one molecule of 3-phosphoglycerate and 2-phosphoglycolate. The turnover rates of Rubiscos are extremely low, and the oxygenase activity further decreases their overall performance in CO2 fixation (1, 11).
Rubiscos that function in the CBB pathway are classified into the type I and type II enzymes. Type I Rubiscos are composed of large and small subunits (L8S8) and are the predominant Rubiscos found in most autotrophic organisms dependent on the CBB pathway. In higher eukaryotes such as plants and green algae, the large subunit that harbors the catalytic center is encoded by the plastomic rbcL gene, whereas the small subunit is encoded by the nuclear rbcS gene (7, 36). In contrast, type II Rubiscos, composed solely of two large subunits, are found in only some photoautotrophic and chemoautotrophic bacteria (36).
Improvements in Rubisco function are expected to have a large impact on various fields of agriculture. However, structure-function studies on the eukaryotic type I enzymes have been hampered by the fact that functional expression of these proteins in conventional host cells such as Escherichia coli is not possible (6, 33). On the other hand, bacterial type I enzymes from cyanobacteria and type II Rubiscos can be functionally expressed in Escherichia coli (8, 10, 29), and extensive studies have been carried out in order to understand the structural elements that control the specificity and activity levels of these enzymes (22).
Although archaea do not seem to harbor a functional CBB pathway, structurally related proteins with Rubisco activity have been found and are classified as the type III Rubiscos (3, 37). Activity has been confirmed with the enzymes from Thermococcus kodakaraensis KOD1 (3), Methanocaldococcus jannaschii (37), Archaeoglobus fulgidus (4, 16), and several other methanogenic archaea (4). The enzyme from T. kodakaraensis (RbcTk) is composed only of large subunits and displays extreme thermostability with high carboxylase activity at high temperatures (3, 19). The enzyme exhibits a novel quaternary structure and was found to be a toroid-shaped pentagonal decamer comprised of five L2 dimers (14). It has recently been shown that the type III RbcTk is involved in AMP metabolism in T. kodakaraensis (25).
At ambient temperatures, the carboxylase activity of RbcTk can be expected to be lower than those of mesophilic Rubiscos. However, we have found that RbcTk was able to support both photoautotrophic and photoheterotrophic growth of a Rubisco-deficient mutant strain (strain Δ3) of the mesophilic purple nonsulfur bacterium Rhodopseudomonas palustris No.7 (38). This encouraged us to examine the enzymatic properties of RbcTk at mesophilic temperatures and the possibilities of improving the catalytic performance of the enzyme at these temperatures via protein engineering. This would provide an entirely different approach to obtaining a Rubisco protein with improved or desired enzymatic properties. RbcTk has the advantage that it can be readily expressed at high levels in an active form in E. coli, and as the protein is comprised only of large subunits (19), it may also be easier to express the protein in a functional form in other host environments such as the eukaryotic chloroplast. The high (thermo)stability of the enzyme (18) suggests that the enzyme provides a stable protein scaffold that should be able to tolerate higher degrees of mutations at ambient temperatures than the mesophilic Rubiscos. These properties, along with the elucidated three-dimensional structure of the protein (14), should allow us to introduce robust mutations into RbcTk and possibly improve its performance at mesophilic temperatures.
In this study, we aimed to improve the function of RbcTk and gain insight into the structure-function relationship of the enzyme at ambient temperatures. We have performed site-directed mutagenesis on RbcTk, based on a comparison between sequences of RbcTk and type I/II Rubiscos. The catalytic properties of purified recombinant proteins were examined in vitro, and R. palustris Δ3 was utilized as a host strain for evaluating the performance of these mutant proteins in vivo.
MATERIALS AND METHODS
Strains, media, and growth conditions.
The rbcTk gene was isolated from T. kodakaraensis KOD1, a hyperthermophilic archaeon isolated from Kodakara Island, Kagoshima, Japan (2, 20). R. palustris Δ3 is a derivative of R. palustris No.7 disrupted in its three Rubisco genes, rbc I-1, rbc I-2, and rbc II (38). Gene manipulation and plasmid construction were performed with E. coli DH5α, and E. coli BL21(DE3)CodonPlus RIL (Stratagene, La Jolla, CA) was used for gene expression. E. coli cells were grown aerobically at 37°C in Luria-Bertani (LB) medium with ampicillin (100 μg/ml). R. palustris cells were first cultivated aerobically in the dark at 30°C in LB medium containing 0.3% NaCl and zeocin (400 μg/ml) (heterotrophic growth) for 2 days. Cells were collected, and the pellet was washed and resuspended in a basal salt medium (5) but without yeast extract (BS medium) containing 5 mM NaHCO3 and 50 mM ethanol. Cells were inoculated into BS medium containing 5 mM NaHCO3 and 50 mM ethanol at an initial optical density at 660 nm of 0.05, and photoheterotrophic growth was carried out with a light intensity of 3,200 lx at 25°C. Under photoheterotrophic conditions, cells were grown in a sealed glass vial filled with medium so that the headspace was minimal. Specific growth rates were determined in the exponential growth phase.
DNA manipulation.
DNA manipulation was carried out by using standard methods (24). Plasmid DNA was isolated with a QIAGEN Plasmid Mini kit (QIAGEN, Hilden, Germany). E. coli DH5α was transformed by the CaCl2 method. R. palustris Δ3 was transformed by electroporation. Restriction enzymes were purchased from Toyobo (Osaka, Japan) or Takara Bio (Otsu, Japan). DNA ligation was performed with a DNA ligation kit (Toyobo). KOD Plus (Toyobo) was used as a polymerase for PCR, and a GFX PCR DNA and gel band purification kit (GE Healthcare Bio-Sciences, Piscataway, NJ) was used to recover DNA fragments from agarose gels after electrophoresis. DNA sequencing was performed with a BigDye Terminator Cycle sequencing kit (version 3.1) and a model 3100 capillary DNA sequencer (Applied Biosystems, Foster City, CA).
Mutagenesis and plasmid construction.
For expression plasmids, inverse PCR was performed with pET-21a(+)/rbcTk (3) or plasmids harboring rbcTk mutants (SP2 and GP1) as template DNA. Primers for inverse PCR were constructed so that the regions to be exchanged were removed. 5′-Phosphorylated oligonucleotides harboring the mutant sequences were annealed and ligated with the DNA fragments amplified by inverse PCR. Point mutagenesis was carried out using a QuickChange XL site-directed mutagenesis kit (Stratagene). The DNA manipulation strategy and primers are described in Fig. S1 and Tables S1 to S3 in the supplemental material.
Plasmids used to integrate each mutant gene into the chromosome of R. palustris Δ3 were constructed using pSZMRbc (38). Expression plasmids harboring the mutant rbcTk genes were digested with MroI, and the fragments containing the mutant sequences were ligated with pSZMRbc digested with MroI.
Gene expression and initial examination of carboxylase activity.
RbcTk and its mutants were expressed as described previously (18). E. coli BL21(DE3)CodonPlus RIL cells were collected (10 min, 5,000 × g, 4°C) and washed with 100 mM Bicine-NaOH (pH 8.3)-10 mM MgCl2 (buffer A). The pellet was resuspended in the same buffer and disrupted by sonication on ice. Cell debris was removed by centrifugation (5 min, 5,000 × g, 4°C). Cell extracts were subjected to heat treatment (85°C, 30 min) and centrifugation (30 min, 20,000 × g, 4°C). Further purification of wild-type RbcTk and mutant proteins SP3, SP4, SP6, and SP4-N333F was performed as described previously (18). The protein concentration was determined with a protein assay kit (Bio-Rad, Hercules, CA) by using bovine serum albumin as a standard. Carboxylase activity was measured as described elsewhere previously (26), with slight modifications (38). All assays were performed at least twice.
Western blot analysis.
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis was performed with 12.5% gels. Proteins were blotted from the polyacrylamide gel to polyvinylidene difluoride membranes (Millipore, Bedford, MA). Antisera containing polyclonal rabbit antibodies against purified recombinant RbcTk were used to detect recombinant RbcTk. Samples were prepared from cell extracts of exponentially growing R. palustris cells. Cells were harvested by centrifugation and washed with buffer A. The pellet was resuspended in the same buffer and disrupted by sonication. Cell debris was removed by centrifugation (5 min, 5,000 × g, 4°C) followed by ultracentrifugation (20 min, 110,000 × g, 4°C).
Kinetic examination.
All buffers were autoclaved (121°C, 20 min), cooled, and bubbled under an N2 atmosphere free of CO2 and O2. Prior to use, enzymes were treated with CO2- and O2-free buffer (10 mM Bicine-NaOH [pH 8.3]) on a PD-10 gel filtration column (GE Healthcare Bio-Sciences). Enzymes were concentrated and preincubated for 4 h on ice in a solution containing 10 mM NaHCO3, 10 mM MgCl2, and 10 mM Bicine-NaOH (pH 8.3) for enzyme activation. The reaction was initiated by combining 230 μl of reaction buffer with 20 μl of activated enzyme after each one was preincubated at 25°C for 5 min. The final reaction mixture contained 10 μg activated enzyme, 20 μM RuBP (Fluka, Buchs, Switzerland), 10 mM MgCl2, 10 mM Bicine-NaOH (pH 8.3), and 0.8, 1.8, 2.8, 3.8, 4.8, 6.8, 8.8, or 10.8 mM NaHCO3. Reactions carried out at 25°C were stopped after 30 s and 60 s with 250 μl of 40 mM HCl.
KO2 and the substrate specificity factor (τ) values, VCO2KO2/VO2KCO2, were determined at fixed NaHCO3 concentrations. O2 saturating buffer (saturating [O2] = 1.23 mM, employed for 100% O2-flushed reactions) (30) was prepared by O2 bubbling of CO2- and O2-free buffer for 4 h and added to the reaction mixture. The reaction was initiated with the addition of 230 μl reaction buffer to 20 μl of activated enzyme after each one was preincubated at 25°C for 5 min. The final reaction mixture contained 10 μg (for wild-type RbcTk, SP4, SP6, and SP4-N333F) or 15 μg (for SP3) activated enzyme, 10 μM RuBP, 10 mM MgCl2, 10 mM Bicine-NaOH (pH 8.3), a fixed concentration of NaHCO3, and 0.886 mM O2 or 0 mM O2. The NaHCO3 concentration was 8.8 mM (for wild-type RbcTk), 5.8 mM (SP4, SP6, and SP4-N333F), or 4.8 mM (SP3). Reactions at 25°C were stopped after 30 s, 60 s, and 90 s with 250 μl of 40 mM HCl. The KO2 was calculated using the relationship 1/(R − 1) = KO2/[O2] + KO2[CO2]/KCO2[O2] (17), where R is the ratio of carboxylase activities at O2 concentrations of 0 and 0.886 mM.
Reaction mixtures were analyzed as described elsewhere previously (32), with the modifications noted below. In order to remove Mg2+, reaction mixtures were mixed with Dowex 50 resin (H+ form) (Sigma-Aldrich, St. Louis, MO) washed with 1 M NaOH, followed by 1 M HCl and then MilliQ water. Rubisco and resin were removed with an Ultrafree-MC filter unit (Millipore) by centrifugation (5,000 × g, 4°C), and the filtrate was injected to a DX 500 anion-exchange chromatographic system with an AS11 column (Dionex, Sunnyvale, CA). The developing solvent was 18 mM NaOH.
For the determination of KRuBP, the reaction was initiated with the addition of 40 μl of activated enzyme to 460 μl reaction mixture after each one was preincubated at 25°C for 5 min. The 500-μl reaction mixture contained 2 μg activated enzyme, 5, 10, 15, 20, or 50 μM RuBP, 10 mM MgCl2, 2.8 mM NaHCO3, and 1.8 mM Bicine-NaOH (pH 8.3). Reactions at 25°C were stopped after 30 s and 60 s with Dowex 50 resin (H+ form) treated as described above. Rubisco and resin were removed with an Ultrafree-MC filter and analyzed with the DX 500 anion-exchange chromatographic system.
Effects of temperatures on enzyme stabilities.
Thermostabilities of RbcTk, SP4, SP6, and SP4-N333F were analyzed by measuring the residual carboxylase activities of the proteins after incubation at 90°C or 100°C in buffer A. Denatured proteins were removed by centrifugation (20 min, 15,000 × g, 4°C) before testing. Residual activities were measured with the enzymatic assay methods described above.
RESULTS
Sequence comparison of various Rubiscos and design of RbcTk mutant proteins.
Loop 6 covers the α/β barrel of Rubisco and acts as a lid during Rubisco catalysis. Loop 6 is commonly found in type I to type III Rubiscos and harbors the active-site residue lysine. Structural studies have suggested that the flexible loop 6 in the closed conformation serves in stabilizing the reaction intermediates during catalysis (15, 21). As differences in sequence were observed in this region among the enzymes from higher plants, algae, and cyanobacteria, loop 6 and the adjacent α-helix 6 region have been major targets for site-directed mutagenesis. These studies have confirmed that loop 6/α-helix 6 are involved in defining the specificity factor and turnover rate of type I and type II Rubiscos (22). An alignment of the sequences of this region in enzymes from spinach (type I), the purple nonsulfur bacterium Rhodospirillum rubrum (type II), the red alga Galdieria partita (type I), and RbcTk (type III) is shown in Fig. 1. As the corresponding region in RbcTk displays a further variation among the Rubiscos, we focused on loop 6/α-helix 6 of RbcTk for introducing specific mutations and examining their effects on Rubisco activity at mesophilic temperatures. In addition, increasing the flexibility of the protein at or in the vicinity of the catalytic center may lead to increased turnover of the enzyme at lower temperatures.
FIG. 1.
Mutant design of RbcTk. Shown is an alignment of the loop 6 and α-helix 6 regions of Rubisco proteins from T. kodakaraensis KOD1 (Tk) (type III; GenBank accession number BAD86479), spinach (Sp) (type I; accession number P00875) (red), G. partita (Gp) (type I; accession number BAA75796) (blue), and R. rubrum (Rr) (type II; accession number P04718) (green). Active-site residues are indicated with asterisks. Residues identical in all four enzymes are shaded. Gaps in the alignment are indicated with hyphens. Secondary structural elements are shown above the alignment. (A and B) Sequences of mutant RbcTk proteins produced in this study. Exchanged residues are indicated with the colors of the sequence on which the mutations were based. The positions where single residues were deleted in ΔT317 and ΔA318 are underlined. (B) Sequences of mutant proteins based on SP4.
Ten initial mutant proteins were constructed by replacing sequences of RbcTk with sequences from type I/II Rubiscos (Fig. 1A). Mutants SP1 to SP7 are based on sequence comparisons with the type I spinach Rubisco and results of previously reported mutagenesis studies. Spinach is a terrestrial organism, and its Rubisco exhibits a high CO2/O2 specificity factor (12, 23). A large portion of the residues in loop 6 are highly conserved among the type I to type III Rubiscos. SP1 and SP2 harbor mutations in the nonconserved residues of loop 6. Within α-helix 6, SP3 contains a single amino acid substitution from valine to threonine based on the finding that a T342V mutant clearly decreased the specificity factor and the specific activity of a type I Rubisco from cyanobacteria (23). SP4 has substitutions in five consecutive amino acid residues, taking into account that a DKAS338-341ERDI mutation in a cyanobacterial type I Rubisco led to an increase in the specificity factor (9, 13), in addition to the results for the T342V mutation described above. As RbcTk was presumed to provide a stable scaffold, we introduced mutations to a greater extent in SP5 to SP7. SP5 has a 6-amino-acid substitution in α-helix 6, while SP6 has 11 substitutions in α-helix 6. In SP7, the entire loop 6/α-helix 6 region was replaced with the corresponding region of spinach Rubisco. GP1 and GP2 mutants were designed by taking into consideration the sequence of the type I Rubisco from G. partita. G. partita is a thermophilic red alga, and the enzyme displays the highest specificity reported among Rubiscos (31). The α-helix 6 region of RbcTk was replaced with that of the G. partita Rubisco in GP1, and the entire loop 6/α-helix 6 region was exchanged in GP2. RR1 is based on the sequence from a type II Rubisco from R. rubrum. As the R. rubrum enzyme exhibits a low specificity factor compared with type I Rubisco (12, 13), RR1 was constructed mainly to examine the significance in the length of loop 6.
Carboxylase activities of recombinant mutant RbcTk proteins.
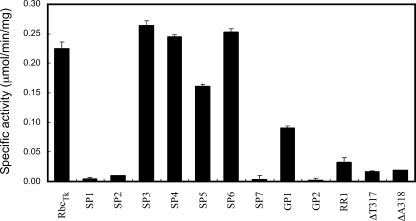
Plasmids were constructed to express the 10 mutant protein genes in E. coli. Cells were disrupted by sonication, and the supernatants were treated with heat at 85°C for 15 min to remove proteins from the mesophilic host cells. After centrifugation, soluble proteins in the supernatants were examined by sodium dodecyl sulfate-polyacrylamide gel electrophoresis, and it was confirmed that all mutant proteins were present in a soluble form and that the proteins were apparently homogeneous. In order to select enzymes for further detailed analysis and to identify mutant enzymes whose activities were completely abolished, we carried out an initial examination of activity for these proteins using the supernatants obtained after heat treatment. At 25°C, activity was clearly detected in the SP3, SP4, SP5, SP6, GP1, and RR1 proteins (Fig. 2). In particular, SP3, SP4, and SP6 exhibited activity levels comparable to the activity observed in RbcTk. On the other hand, SP1, SP2, SP7, and GP2 displayed dramatic decreases in carboxylase activity, indicating that the length of loop 6 is important for the activity of RbcTk. To examine whether this was the case, we constructed two additional mutant proteins with single amino acid deletions in T317 (ΔT317) and A318 (ΔA318). Dramatic decreases in activities were observed in both mutant proteins (Fig. 2), further supporting the importance of the length of loop 6 in RbcTk activity.
FIG. 2.
Activity levels of partially purified wild-type and mutant RbcTk proteins. Carboxylase activities of partially purified wild-type and mutant RbcTk proteins are indicated. Activity measurements were performed at 25°C. Measurements were performed at least in duplicate.
Growth characteristics of R. palustris Δ3 strains harboring mutant RbcTk proteins.
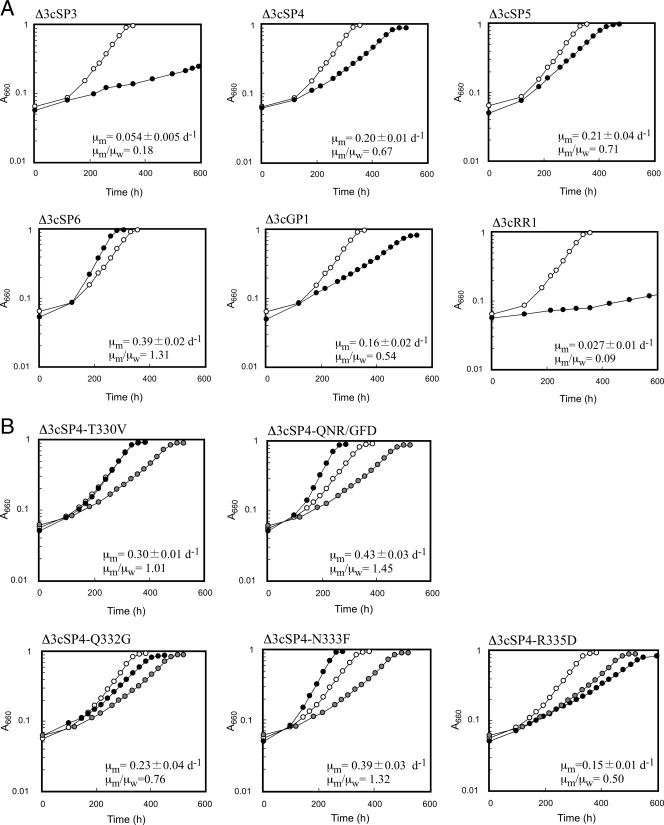
The six mutant proteins with relevant levels of activity (SP3, SP4, SP5, SP6, GP1, and RR1) were examined for their ability to support the growth of the Rubisco-deficient mutant strain R. palustris Δ3. The genes were individually introduced into R. palustris Δ3 cells using plasmid pSMZRbc, designed for single-crossover insertion into the R. palustris Δ3 chromosome (38). Transformants were selected for their resistance to zeocin, and genotypes were confirmed by PCR analyses (data not shown). R. palustris Δ3 strains with the mutated rbcTk genes were designated strains Δ3cSP3, Δ3cSP4, Δ3cSP5, Δ3cSP6, Δ3cGP1, and Δ3cRR1, corresponding to the mutant proteins that they produce. Δ3cRbcTk, harboring the wild-type rbcTk gene on the chromosome, and the six strains with mutant proteins were cultivated aerobically in LB medium for 2 days, and the cells were washed and grown under photoheterotrophic conditions. We have previously shown that Rubisco activity is essential for growth under these conditions and that specific growth rates correlate well with the levels of Rubisco activity found in the cell extracts (38). We found that the six strains with mutant RbcTk proteins could grow under photoheterotrophic conditions. The strains exhibited distinct specific growth rates depending on the protein that they harbored. Three to seven independent cultures were performed for each strain. Δ3cSP6 cells exhibited the highest specific growth rate (0.39 ± 0.02 day−1), followed by Δ3cRbcTk (0.29 ± 0.02 day−1), Δ3cSP5 (0.21 ± 0.04 day−1), Δ3cSP4 (0.20 ± 0.01 day−1), Δ3cGP1 (0.16 ± 0.02 day−1), Δ3cSP3 (0.054 ± 0.005 day−1), and Δ3cRR1 (0.027 ± 0.01 day−1) (Fig. 3A). The levels of cell yield after full growth in these batch cultures were equivalent (not shown). Δ3cSP6 displayed a 31% increase in the specific growth rate compared to that of Δ3cRbcTk. The other recombinant strains grew at similar or lower specific growth rates compared to that of Δ3cRbcTk. To confirm the expression levels of the RbcTk protein in each recombinant strain, Western blot analysis was carried out on the cell extracts of exponentially growing cells. The bands corresponding to the RbcTk proteins were equivalent in intensity (not shown), suggesting that the differences in growth rates were a result of differences in the performance of the individual proteins.
FIG. 3.
Growth of R. palustris Δ3 recombinant strains harboring wild-type and mutant RbcTk proteins. Representative growth curves of Δ3cRbcTk (open circles) and strains harboring mutant proteins (closed circles) under photoheterotrophic conditions are shown. Heterotrophically grown cells were washed and inoculated into photoheterotrophic medium. The average specific growth rate of seven independent growth experiments of Δ3cRbcTk (μw) was 0.30 ± 0.02 day−1. The average specific growth rate of each strain harboring a mutant RbcTk (μm) (three to five measurements) was compared with that of μw and is indicated in each panel. (A) Growth of strains harboring mutant Rubiscos that displayed relevant levels of activity shown in Fig. 2. (B) Growth of strains harboring mutant Rubisco proteins based on SP4. A representative growth curve for Δ3cSP4 is also indicated with gray circles as a comparison.
Kinetic analysis of the purified recombinant enzymes.
We examined and compared the kinetic parameters of RbcTk and mutant SP6, which led to a 31% increase in the specific growth rate. We also examined two other proteins with levels of activity similar to those of RbcTk: SP4, which led to lower growth rates than RbcTk, and SP3, whose performance in vivo was dramatically lower than that of the wild-type protein. E. coli cell extracts containing the recombinant proteins (RbcTk and mutants SP3, SP4, and SP6) were subjected to heat treatment and anion-exchange and gel filtration chromatography and purified to apparent homogeneity. Kinetic analyses were performed at 25°C.
The maximum velocity of the carboxylase reaction (VCO2) for wild-type RbcTk was 0.30 μmol min−1 mg−1, much lower than those of previously examined Rubiscos from mesophilic organisms (Table 1). The KCO2 value of RbcTk was 52.3 μM, a value lower than those observed for the type I Rubiscos from cyanobacteria but higher than those of plant Rubiscos. The substrate specificity factor τ of RbcTk was 11.2 at 25°C, which is a considerably low value compared with those of spinach, Synechococcus, and other type I Rubiscos. On the other hand, RbcTk exhibited an extremely low KRuBP value of <0.5 μM, which was the lowest among those previously reported. Accurate KRuBP values could not be obtained using an anion-exchange chromatographic system with an AS11 column.
TABLE 1.
Kinetic properties of purified RbcTk and its mutant proteins at 25°C
| Protein | Growth ratea (day−1) | VCO2 (μmol/min/mg) ± SD | kcat(CO2) (s−1·site−1) | KCO2 (μM) ± SD | kcat(CO2)/KCO2 ratio | VO2 (μmol/min/mg) | kcat(O2) (s−1·site−1) | KO2 (μM) ± SD | kcat(O2)/KO2 ratio | τ (VCO2KO2/VO2KCO2) | KRuBP (μM) ± SD |
|---|---|---|---|---|---|---|---|---|---|---|---|
| RbcTk proteins | |||||||||||
| RbcTk | 0.30 | 0.30 ± 0.01 | 0.25 | 52 ± 6 | 0.0047 | 0.46 | 0.38 | 900 ± 90 | 0.00042 | 11 ± 1 | <0.5 |
| SP3 | 0.054 | 0.28 ± 0.01 | 0.23 | 74 ± 8 | 0.0031 | 0.06 | 0.05 | 280 ± 20 | 0.00017 | 18 ± 1 | <0.5 |
| SP4 | 0.20 | 0.30 ± 0.02 | 0.25 | 50 ± 8 | 0.0050 | 0.17 | 0.14 | 300 ± 10 | 0.00047 | 11 ± 0 | <0.5 |
| SP6 | 0.39 | 0.39 ± 0.02 | 0.33 | 59 ± 6 | 0.0055 | 0.29 | 0.24 | 460 ± 40 | 0.00051 | 11 ± 2 | <0.5 |
| SP4-N333F | 0.39 | 0.57 ± 0.04 | 0.47 | 78 ± 10 | 0.0060 | 0.24 | 0.47 | 190 ± 20 | 0.0010 | 6.0 ± 0.4 | <0.5 |
| Reference proteins | |||||||||||
| Spinach (31) | 2.3 | 2.6 | 14.3 | 0.182 | 93.8 ± 0.8 | ||||||
| R. rubrum (27) | 6.2 ± 0.4 | 5.2 | 150 ± 30 | 0.035 | 1.57 | 1.32 | 380 ± 50 | 0.0035 | 10 ± 0.5 | ||
| Synechococcus (23) | 3.24 ± 0.07 | 3.71 | 167 ± 2 | 0.022 | 0.25 | 0.29 | 529 ± 14 | 0.00053 | 41.0 ± 4.4 | 19.2 ± 3.6 |
Specific growth rates of recombinant strains which harbor the corresponding proteins.
We clearly observed an increase in turnover number in the SP6 protein (30% increase in VCO2), which resulted in a 17% increase in kcat(CO2)/KCO2. In the case of SP4, the kinetic parameters for the carboxylase activity were similar to those of RbcTk. In the case of SP3, the protein displayed a decrease in VCO2, along with a relatively large increase in KCO2, resulting in a 35% decrease in kcat(CO2)/KCO2 (Table 1).
Further analyses of the mutant proteins SP4 and SP6.
The single mutation V330T in SP3 had a relatively large negative effect on RbcTk activity. We further examined the effect of this single mutation by exchanging T330 of SP4 back to valine (SP4-T330V). Growth rates of an R. palustris Δ3 strain harboring the SP4-T330V gene (Δ3cSP4-T330V) were higher than those of strain Δ3cSP4 and equivalent to those of R. palustris Δ3 harboring wild-type RbcTk (Fig. 3B), confirming the negative effects of the V330T mutation.
As both in vitro (kinetics) and in vivo (growth rates) analyses indicated that the SP6 mutant protein displayed improved catalytic properties, we constructed several mutant proteins in order to examine which residues were responsible for this effect. However, the improved performance of SP6 was not expected to be a simple additive sum of effects brought about by individual residue replacements. Neither SP4 nor SP5, whose mutations, when combined, correspond to SP6, exhibited improved enzyme performance. As we had examined SP4 and SP6 both kinetically and in vivo, we focused here on the residues responsible for the differences in performance observed between SP4 and SP6.
Six residues are different between SP4 and SP6 (positions 331 to 336). Among these, we presumed that the differences in residues at positions 332 (SP4:Gln/SP6:Gly), 333 (SP4:Asn/SP6:Phe), and 335 (SP4:Arg/SP6:Asp) would have greater effects than the differences at positions 331 (Ile/Leu), 334 (Ala/Val), and 336 (Ile/Leu). We therefore constructed a mutant protein (SP4-QNR/GFD) where residues of SP4 at positions 332, 333, and 335 were replaced by the corresponding residues of SP6. The SP4-QNR/GFD gene was introduced into R. palustris Δ3 cells (Δ3cSP4-QNR/GFD), and photoheterotrophic growth rates were examined (Fig. 3B). The specific growth rate of Δ3cSP4-QNR/GFD (0.43 day−1) displayed a significant increase compared to that of SP4 (0.20 day−1) and was even higher than the specific growth rate of SP6 (0.39 day−1). This strongly suggests that the residue(s) responsible for the differences in activity between SP4 and SP6 is included in these three residues. We thus examined the effects of changing each individual residue (SP4-Q332G, SP4-N333F, and SP4-R335D). R. palustris Δ3 cells harboring the SP4-R335D gene exhibited a specific growth rate (0.15 day−1) even lower than that of the SP4 strain (0.20 day−1), indicating that D335 of SP6 does not contribute to the increase in SP6 activity compared to that of SP4 and, rather, that it has a negative effect. Growth rates of cells producing SP4-Q332G were higher than those of Δ3cSP4 cells, indicating that the Q332G mutation does have a positive effect, but growth rates were still lower than those of Δ3cSP6 and Δ3cRbcTk. The specific growth rate of cells harboring the SP4-N333F gene (Δ3cSP4-N333F) displayed a dramatic increase (0.39 day−1) compared to that of Δ3cSP4, equivalent to that of Δ3cSP6 (0.39 day−1), suggesting that F333 is the residue that is most responsible for the differences in enzyme performance between SP4 and SP6 (Fig. 3B). The SP4-N333F protein was thus purified and subjected to kinetic analyses. As in the case of SP6, the most significant change in properties between the SP4-N333F protein and wild-type RbcTk was in the turnover numbers of their carboxylase activities, with a 90% increase in VCO2 observed in SP4-N333F, resulting in a 28% increase in kcat(CO2)/KCO2 (Table 1).
Effects of temperatures on enzyme stabilities.
We have shown that by replacing residues of the thermophilic RbcTk protein with residues found in mesophilic Rubiscos, it was possible to enhance the performance of RbcTk at ambient temperatures. In general, thermostable proteins are presumed to obtain their thermostability by increasing their conformational rigidity compared to enzymes from mesophiles (35, 39). This in turn, in many cases, results in lower activity levels of the thermostable enzymes than those of their mesophilic counterparts at ambient temperatures (34). Increasing the flexibility of these enzymes can be expected to be one method to enhance the activity, particularly the turnover rates, of these enzymes at low temperatures. On the other hand, adding flexibility to an enzyme can also be considered to have a negative effect on the thermostability of the enzyme. To examine whether this was the case in our RbcTk mutant proteins, we measured and compared the thermostabilities of wild-type RbcTk, SP4, SP6, and SP4-N333F. At both 90°C and 100°C, there was a tendency that proteins with higher numbers of residues from the mesophilic spinach Rubisco were less stable (Table 2). Moreover, we found that SP6 and SP4-N333F, whose kcat values were considerably higher than those of RbcTk and SP4, exhibited large decreases in thermostability. This raises the possibilities that in SP6 and SP4-N333F, the increase in turnover rates may be brought about by the increased flexibility of the α-helix 6 region at the expense of decreasing the overall thermostability of the enzyme.
TABLE 2.
Thermostability of RbcTk and its mutant proteins
| Temp (°C) | Half-life (min) ± SD
|
|||
|---|---|---|---|---|
| RbcTk | SP4 | SP6 | SP4-N333F | |
| 90 | 220 ± 10 | 190 ± 19 | 120 ± 8 | 160 ± 13 |
| 100 | 48 ± 2 | 33 ± 3 | 2.5 ± 0.1 | 17 ± 1 |
DISCUSSION
In order to examine the possibilities of improving the function of RbcTk at ambient temperatures, we performed an initial site-directed mutagenesis study on the enzyme. Focusing on loop 6 and α-helix 6, we constructed mutant proteins based on the sequences of mesophilic or moderately thermophilic type I and type II enzymes. Although a number of mutant proteins had over 10 residues exchanged, we found that all proteins were produced in a soluble form in E. coli. This confirmed our assumption that the extremely thermostable scaffold of RbcTk is able to tolerate large extents of mutagenesis and will provide an advantage in further studies taking a random mutagenesis approach.
The kinetic analysis of wild-type RbcTk indicated that the enzyme displays a very low turnover number at 25°C, which can be expected, as the enzyme is derived from a hyperthermophile. The affinity for CO2 was higher than that of the type I enzyme from Synechococcus but lower than the values for the spinach enzyme. A notable feature was the extremely low Km value for RuBP. This trait is most likely due to the necessity of the enzyme to rapidly bind to RuBP in T. kodakaraensis at high temperatures, as RuBP is a thermolabile compound.
The loop 6 and α-helix 6 regions have been demonstrated to play an important role in the turnover and specificity of type I and type II Rubiscos (22). Our results have clarified that these regions are also important for the activity of the type III RbcTk. The length of loop 6 is vital; a deletion of a single amino acid residue results in an almost complete loss of activity in mutants SP3, SP4, SP7, GP1, ΔT317, and ΔA318 regardless of the sequences in the α-helix 6 region. We also found that residue replacements in loop 6 (mutant RR1) also dramatically affect specific activity levels. As for α-helix 6, a replacement with the corresponding region from G. partita led to an over 50% reduction in activity levels, while replacement with the region from spinach had a positive effect. The results indicate that the α-helix 6 region also plays a key role in RbcTk catalysis.
Mutant Rubisco proteins that exhibited activity in vitro were introduced into R. palustris Δ3, and growth under photoheterotrophic conditions was analyzed. As the growth experiments were performed in the presence of 5 mM NaHCO3 and with little headspace in the sealed vials, the turnover rate of the carboxylase activity (VCO2) was expected to have a large effect on the specific growth rates. The kcat(CO2)/KCO2 values may also be relevant, depending on the capacity of NaHCO3 uptake in R. palustris and the intracellular levels of carbonic anhydrase activity. Growth examinations of the initial six recombinant strains (Fig. 3A) revealed that Δ3cSP6 grew at a higher rate than Δ3cRbcTk (31% increase). This correlates well with the fact that the SP6 protein exhibited higher VCO2 and kcat(CO2)/KCO2 values than did wild-type RbcTk (Table 1). The increases in these values most likely override the slight decrease in the τ value. As for Δ3cSP3, the strain exhibited much slower growth than did Δ3cRbcTk (82% decrease). Accordingly, the VCO2 and kcat(CO2)/KCO2 values displayed 9% and 34% decreases, respectively. The increase in the τ value, due mainly to the decrease in oxygenase activity (and not an increase in carboxylase activity), was not reflected under our growth conditions. In the case of SP4, it was difficult to determine what properties of the protein were responsible for the 33% decrease in the specific growth rate, as the kinetic properties in vitro were apparently equivalent to those of wild-type RbcTk. The SP4-N333F mutant protein allowed us to determine that among the six residues exchanged between SP4 and SP6, the presence of phenylalanine at position 333 contributed most to the improved performance in SP6. This position most likely plays an important role in Rubisco activity. The effects of a mutation in the corresponding position (F342V) in a type I Rubisco from Synechococcus have been examined both in vitro and in vivo. Although the kinetic parameters displayed only an improvement in the affinity for RuBP, Rubisco-deficient Rhodobacter capsulatus cells harboring the F342V enzyme displayed significantly higher rates of photoautotrophic growth than cells with the wild-type enzyme (28).
SP6 and SP4-N333F, two mutant proteins that supported higher growth rates in R. palustris Δ3, both displayed increases in their turnover numbers compared to those of wild-type RbcTk. In addition to the effect of specific residue replacements, the increase in turnover number may also be due to an increase in flexibility in these regions, as a clear decrease in the thermostability of these proteins was observed. Further mutations aimed to decrease the rigidity of RbcTk near its active site, such as disrupting residue interactions found in RbcTk but not in mesophilic Rubiscos, may lead to further increases in the catalytic activity of RbcTk at mesophilic temperatures.
Supplementary Material
Acknowledgments
This study was partly supported by the Research Institute of Innovative Technology for the Earth (RITE) (to T.I.).
Footnotes
Published ahead of print on 3 August 2007.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Andrews, T. J., and G. H. Lorimer. 1987. Rubisco: structure, mechanisms, and prospects for improvement, p. 131-218. In M. D. Hatch and N. K. Boardman (ed.), The biochemistry of plants. Academic Press, San Diego, CA.
- 2.Atomi, H., T. Fukui, T. Kanai, M. Morikawa, and T. Imanaka. 2004. Description of Thermococcus kodakaraensis sp. nov., a well studied hyperthermophilic archaeon previously reported as Pyrococcus sp. KOD1. Archaea 1:263-267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ezaki, S., N. Maeda, T. Kishimoto, H. Atomi, and T. Imanaka. 1999. Presence of a structurally novel type ribulose-bisphosphate carboxylase/oxygenase in the hyperthermophilic archaeon, Pyrococcus kodakaraensis KOD1. J. Biol. Chem. 274:5078-5082. [DOI] [PubMed] [Google Scholar]
- 4.Finn, M. W., and F. R. Tabita. 2003. Synthesis of catalytically active form III ribulose 1,5-bisphosphate carboxylase/oxygenase in archaea. J. Bacteriol. 185:3049-3059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fujii, T., A. Nakazawa, N. Sumi, H. Tani, A. Ando, and M. Yabuki. 1983. Utilization of alcohols by Rhodopseudomonas sp. no. 7 isolated from n-propanol-enrichment cultures. Agric. Biol. Chem. 47:2747-2753. [Google Scholar]
- 6.Gatenby, A. A. 1988. Synthesis and assembly of bacterial and higher plant Rubisco subunits in Escherichia coli. Photosynth. Res. 17:145-157. [DOI] [PubMed] [Google Scholar]
- 7.Gatenby, A. A., and R. J. Ellis. 1990. Chaperone function: the assembly of ribulose bisphosphate carboxylase-oxygenase. Annu. Rev. Cell Biol. 6:125-149. [DOI] [PubMed] [Google Scholar]
- 8.Gatenby, A. A., S. M. van der Vies, and D. Bradley. 1985. Assembly in E. coli of a functional multi-subunit ribulose bisphosphate carboxylase from a blue-green alga. Nature 314:617-620. [Google Scholar]
- 9.Gutteridge, S., D. F. Rhoades, and C. Herrmann. 1993. Site-specific mutations in a loop region of the C-terminal domain of the large subunit of ribulose bisphosphate carboxylase/oxygenase that influence substrate partitioning. J. Biol. Chem. 268:7818-7824. [PubMed] [Google Scholar]
- 10.Gutteridge, S., I. Sigal, B. Thomas, R. Arentzen, A. Cordova, and G. Lorimer. 1984. A site-specific mutation within the active site of ribulose-1,5-bisphosphate carboxylase of Rhodospirillum rubrum. EMBO J. 3:2737-2743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Harpel, M. R., E. H. Serpersu, and F. C. Hartman. 1995. Utilization of partial reactions, side reactions, and chemical rescue to analyse site-directed mutants of ribulose 1,5-bisphosphate (RuBP) carboxylase/oxygenase (Rubisco), p. 357-364. In J. W. Crabb (ed.), Techniques in protein chemistry. Academic Press, San Diego, CA.
- 12.Jordan, D. B., and W. L. Ogren. 1981. Species variation in the specificity of ribulose biphosphate carboxylase/oxygenase. Nature 291:513-515. [Google Scholar]
- 13.Kane, H. J., J. Viil, B. Entsch, K. Paul, M. K. Morell, and T. J. Andrews. 1994. An improved method for measuring the CO2/O2 specificity of ribulosebisphosphate carboxylase-oxygenase. Aust. J. Plant Physiol. 21:449-461. [Google Scholar]
- 14.Kitano, K., N. Maeda, T. Fukui, H. Atomi, T. Imanaka, and K. Miki. 2001. Crystal structure of a novel-type archaeal Rubisco with pentagonal symmetry. Structure 9:473-481. [DOI] [PubMed] [Google Scholar]
- 15.Knight, S., I. Andersson, and C.-I. Brändén. 1990. Crystallographic analysis of ribulose 1,5-bisphosphate carboxylase from spinach at 2.4 Å resolution. Subunit interactions and active site. J. Mol. Biol. 215:113-160. [DOI] [PubMed] [Google Scholar]
- 16.Kreel, N. E., and F. R. Tabita. 2007. Substitutions at methionine 295 of Archaeoglobus fulgidus ribulose-1,5-bisphosphate carboxylase/oxygenase affect oxygen binding and CO2/O2 specificity. J. Biol. Chem. 282:1341-1351. [DOI] [PubMed] [Google Scholar]
- 17.Laing, W. A., W. L. Ogren, and R. H. Hageman. 1975. Bicarbonate stabilization of ribulose 1,5-diphosphate carboxylase. Biochemistry 14:2269-2275. [DOI] [PubMed] [Google Scholar]
- 18.Maeda, N., T. Kanai, H. Atomi, and T. Imanaka. 2002. The unique pentagonal structure of an archaeal Rubisco is essential for its high thermostability. J. Biol. Chem. 277:31656-31662. [DOI] [PubMed] [Google Scholar]
- 19.Maeda, N., K. Kitano, T. Fukui, S. Ezaki, H. Atomi, K. Miki, and T. Imanaka. 1999. Ribulose bisphosphate carboxylase/oxygenase from the hyperthermophilic archaeon Pyrococcus kodakaraensis KOD1 is composed solely of large subunits and forms a pentagonal structure. J. Mol. Biol. 293:57-66. [DOI] [PubMed] [Google Scholar]
- 20.Morikawa, M., Y. Izawa, N. Rashid, T. Hoaki, and T. Imanaka. 1994. Purification and characterization of a thermostable thiol protease from a newly isolated hyperthermophilic Pyrococcus sp. Appl. Environ. Microbiol. 60:4559-4566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Newman, J., and S. Gutteridge. 1993. The X-ray structure of Synechococcus ribulose-bisphosphate carboxylase/oxygenase-activated quaternary complex at 2.2-Å resolution. J. Biol. Chem. 268:25876-25886. [PubMed] [Google Scholar]
- 22.Parry, M. A. J., P. J. Andralojc, R. A. C. Mitchell, P. J. Madgwick, and A. J. Keys. 2003. Manipulation of Rubisco: the amount, activity, function and regulation. J. Exp. Bot. 54:1321-1333. [DOI] [PubMed] [Google Scholar]
- 23.Read, B. A., and F. R. Tabita. 1994. High substrate specificity factor ribulose bisphosphate carboxylase/oxygenase from eukaryotic marine algae and properties of recombinant cyanobacterial Rubisco containing “algal” residue modifications. Arch. Biochem. Biophys. 312:210-218. [DOI] [PubMed] [Google Scholar]
- 24.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 25.Sato, T., H. Atomi, and T. Imanaka. 2007. Archaeal type III RuBisCOs function in a pathway for AMP metabolism. Science 315:1003-1006. [DOI] [PubMed] [Google Scholar]
- 26.Schloss, J. V., E. F. Phares, M. V. Long, I. L. Norton, C. D. Stringer, and F. C. Hartman. 1982. Ribulosebisphosphate carboxylase/oxygenase from Rhodospirillum rubrum. Methods Enzymol. 90:522-528. [DOI] [PubMed] [Google Scholar]
- 27.Smith, H. B., F. W. Larimer, and F. C. Hartman. 1990. An engineered change in substrate specificity of ribulosebisphosphate carboxylase/oxygenase. J. Biol. Chem. 265:1243-1245. [PubMed] [Google Scholar]
- 28.Smith, S. A., and F. R. Tabita. 2003. Positive and negative selection of mutant forms of prokaryotic (cyanobacterial) ribulose-1,5-bisphosphate carboxylase/oxygenase. J. Mol. Biol. 331:557-569. [DOI] [PubMed] [Google Scholar]
- 29.Somerville, C. R., and S. C. Somerville. 1984. Cloning and expression of the Rhodospirillum rubrum ribulosebisphosphate carboxylase gene in Escherichia coli. Mol. Gen. Genet. 193:214-219. [Google Scholar]
- 30.Spreitzer, R. J., D. B. Jordan, and W. L. Ogren. 1982. Biochemical and genetic analysis of an RuBP carboxylase/oxygenase-deficient mutant and revertants of Chlamydomonas reinhardii. FEBS Lett. 148:117-121. [Google Scholar]
- 31.Uemura, K., Anwaruzzaman, S. Miyachi, and A. Yokota. 1997. Ribulose-1,5-bisphosphate carboxylase/oxygenase from thermophilic red algae with a strong specificity for CO2 fixation. Biochem. Biophys. Res. Commun. 233:568-571. [DOI] [PubMed] [Google Scholar]
- 32.Uemura, K., Y. Suzuki, T. Shikanai, A. Wadano, R. G. Jensen, W. Chmara, and A. Yokota. 1996. A rapid and sensitive method for determination of relative specificity of RuBisCO from various species by anion-exchange chromatography. Plant Cell Physiol. 37:325-331. [Google Scholar]
- 33.van der Vies, S. M., D. Bradley, and A. A. Gatenby. 1986. Assembly of cyanobacterial and higher plant ribulose bisphosphate carboxylase subunits into functional homologous and heterologous enzyme molecules in Escherichia coli. EMBO J. 5:2439-2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Varley, P. G., and R. H. Pain. 1991. Relation between stability, dynamics and enzyme activity in 3-phosphoglycerate kinases from yeast and Thermus thermophilus. J. Mol. Biol. 220:531-538. [DOI] [PubMed] [Google Scholar]
- 35.Vihinen, M. 1987. Relationship of protein flexibility to thermostability. Protein Eng. 1:477-480. [DOI] [PubMed] [Google Scholar]
- 36.Watson, G. M. F., and F. R. Tabita. 1997. Microbial ribulose 1,5-bisphosphate carboxylase/oxygenase: a molecule for phylogenetic and enzymological investigation. FEMS Microbiol. Lett. 146:13-22. [DOI] [PubMed] [Google Scholar]
- 37.Watson, G. M. F., J.-P. Yu, and F. R. Tabita. 1999. Unusual ribulose 1,5-bisphosphate carboxylase/oxygenase of anoxic Archaea. J. Bacteriol. 181:1569-1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yoshida, S., M. Inui, H. Yukawa, T. Kanao, K. Tomizawa, H. Atomi, and T. Imanaka. 2006. Phototrophic growth of a Rubisco-deficient mesophilic purple nonsulfur bacterium harboring a Type III Rubisco from a hyperthermophilic archaeon. J. Biotechnol. 124:532-544. [DOI] [PubMed] [Google Scholar]
- 39.Závodszky, P., J. Kardos, Á. Svingor, and G. A. Petsko. 1998. Adjustment of conformational flexibility is a key event in the thermal adaptation of proteins. Proc. Natl. Acad. Sci. USA 95:7406-7411. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.