Abstract
Coinfections within hosts present opportunities for horizontal gene transfer between strains and competitive interactions between genotypes and thus can be a critical element of the lifestyles of pathogens. Bartonella spp. are Alphaproteobacteria that parasitize mammalian erythrocytes and endothelial cells. Their vectors are thought to be various biting arthropods, such as fleas, ticks, mites, and lice, and they are commonly cited as agents of various emerging diseases. Coinfections by different Bartonella strains and species can be common in mammals, but little is known about specificity and coinfections in arthropod vectors. We surveyed the rate of mixed infections of Bartonella in flea vectors (Polygenis gwyni) parasitizing cotton rats (Sigmodon hispidus) in which previous surveys indicated high rates of infection. We found that nearly all fleas (20 of 21) harbored one or more strains of Bartonella, with rates of coinfection approaching 90%. A strain previously identified as common in cotton rats was also common in their fleas. However, another common strain in cotton rats was absent from P. gwyni, while a rare cotton rat strain was quite common in P. gwyni. Surprisingly, some samples were also coinfected with a strain phylogenetically related to Bartonella clarridgeiae, which is typically associated with felids and ruminants. Finally, a locus (pap31) that is characteristically borne on phage in Bartonella was successfully sequenced from most samples. However, sequence diversity in pap31 was novel in the P. gwyni samples, relative to other Bartonella previously typed with pap31, emphasizing the likelihood of large reservoirs of cryptic diversity in natural populations of the pathogen.
Most host populations harbor more than one pathogen strain at a given time, leading to mixed infections or “coinfections” in individual hosts (10, 26, 48). Unfortunately, there are gaps in our understanding of within-host pathogen interactions. The problem is particularly acute in vector-borne diseases, where little is known regarding mixed infection interactions in natural populations of the vectors themselves. Rather, with only a few notable exceptions (e.g., reference 25), most population-level or clinical data on mixed infections derive from human studies or other mammalian models. The distinction is crucial because of the role that vectors play in pathogen transmission.
The bacterial pathogen Bartonella sp. has become one of a few model organisms for studying the evolution and ecology of vector-borne diseases (28). This is due to diverse efforts to describe Bartonella biology at multiple levels, from cells and immune systems (12, 13, 14, 30), to populations and communities (31, 32), to species and clades (36, 44). The recent publication of full genome sequences is obviously key (2). Bartonella sp. is a short, gram-negative, fastidious bacterium belonging to the Alphaproteobacteria (1). Closely related to Brucella spp., Bartonella organisms are parasites of mammalian erythrocytes and endothelial cells (12, 13, 14) and are transmitted by blood-feeding insects, such as ticks, fleas, lice, and flies (9, 19, 20, 21, 23, 28). Infection of a host causes chronic bacteremia and creates a reservoir for vectors that can transmit the bacteria to new susceptible hosts. While prolonged bacteremia is normally associated with severe sickness in a susceptible host, Bartonella-caused bacteremia typically remains asymptomatic in the reservoir host. Some bartonellae are known to be transmitted by the bite (anterior station transmission) or in the feces (posterior station transmission) of insect vectors. For example, in humans, Bartonella bacilliformis, which causes Oroya fever (verruga peruana, or Carrion's disease) in Andean South America is transmitted by the bites of infectious sandflies (5), and Bartonella quintana, which causes trench fever in many parts of the world, is transmitted via the feces of infected body lice (21). Fleas infected with Bartonella henselae (the causative agent of cat scratch disease and of related conditions such as bacillary angiomatosis [30]) and other bartonellae appear to transmit these agents via their infectious feces (9, 19, 20). Current phylogenetic information indicates six distinct groups worldwide, of which all but one are found in the United States (44). Host and vector affiliations are complex, and the evidence is against strict one-to-one host specificity (28, 32, 33). A consistent trend is that groups of Bartonella species tend to be restricted to natural groups of mammalian hosts (rodents, cats, dogs, humans, etc.), indicating a diffuse but long-term coevolutionary history.
We surveyed the incidence of mixed Bartonella infections in natural populations of the flea Polygenis gwyni parasitizing the Eastern woodrat (Neotoma floridana) and the hispid cotton rat (Sigmodon hispidus). Previous surveys of mammalian hosts indicated that mixed infections of Bartonella can be common (22). An intensive survey of S. hispidus in the southeastern United States, for example, revealed that this host exhibits a particularly high infection prevalence overall, as well as nonnegligible rates of coinfection (33). There is little comparable information on mixed infection rates in Bartonella vectors (49). However, with mammalian host populations multiply infected with strains that are vectored by insects with generalist host affiliations (e.g., ticks and fleas), the expectation is that rates of mixed infections in competent vectors should be quite high. The relevance of whether or not this is the case not only bears on the basic natural history and disease dynamics of Bartonella but also on the pattern and tempo of disease emergence (5). Bartonella has been described as the consummate “versatile pathogen” (27) for the breadth of its host affiliations and plasticity of its lifestyles (2). Population-level data are necessary supplements to evolutionary inferences about the genus, because retrospective analyses of such events as lateral gene transfer (4) can become forward-looking and predictive when accompanied by real-time data on the ecological context for such events (15, 24, 34, 47, 55).
MATERIALS AND METHODS
Trapping and collection methods.
Cotton rats and Eastern woodrats were trapped in Bulloch and Screven Counties (one site in each county) in southeastern Georgia, using Sherman live traps (H. B. Sherman Traps, Inc., Tallahassee, FL) baited with rolled oats and a trace of peanut butter and set near areas of rodent activity. Trapped animals were lightly anesthetized via intramuscular administration of ketamine hydrochloride and then transferred to a white tray, where they were carefully examined for ectoparasites. Retrieved ectoparasites were transferred to individually labeled cryovials containing 95% ethanol. Fleas were later identified using the methods of Smit (52) and Lewis and Lewis (38). All fleas collected were Polygenis gwyni, which is the species that typically parasitizes the cotton rat in the southern United States (38, 52). This flea species has also been reported previously from the Eastern woodrat (16). Following recovery from anesthesia, all cotton rats and woodrats were released at their capture site. Mammals were live trapped under permit 9172 issued by the Georgia Department of Natural Resources, and animal procedures were approved by the IACUC committee at Georgia Southern University (research protocol no. I06003). Voucher flea specimens have been deposited in the Ectoparasite Collection at Georgia Southern University under accession numbers L-358 and L-1102.
DNA methods.
Whole genomic DNA from P. gwyni was extracted using a DNeasy tissue kit (QIAGEN, Inc.). Each sample was tested for the presence or absence of Bartonella by PCR amplification of an approximately 400-bp amplicon from the citrate synthase gltA gene, using the universal oligonucleotide primers BhCS781.p and BhCs1137.n (33). gltA was chosen because of its high discriminating power for Bartonella (36), the existing coverage in GenBank of the genus using this gene, and its prior use in identifying Bartonella in the flea host, S. hispidus (32). PCR products were visualized by electrophoresis and ethidium bromide staining under UV light on 1.5% agarose gels. Samples yielding successful gltA amplicons were then retested with oligonucleotides papn1 and papn2, designed from the bacteriophage-associated gene pap31 in B. henselae. Both PCR amplifications were carried out at 10-μl volumes, containing 1× Invitrogen 10× buffer, 2.0 mM MgCl2, 100 μM of each deoxynucleoside triphosphate, 5 pmol of each primer, 1 U of Invitrogen DNA Taq polymerase, sterile PCR-grade water, and approximately 5 to 10 ng of whole genomic DNA. Reaction conditions were 1 cycle at 94°C for 2 min and 30 cycles of 94°C for 30 s, 52°C for 30 s, and 72°C for 60 s, followed by 1 cycle at 72°C for 15 min. Products from both reactions were cloned via a pCR2.1-TOPO vector (Invitrogen Life Technologies, Carlsbad, CA) and TOPO TA cloning kit and Top10 competent cells, according to the manufacturer's instructions. Positive clones for both genes were PCR amplified at 50-μl volumes, as above, purified with a QIAGEN PCR purification kit (QIAGEN, Inc.) following the manufacturer's instructions, and sequenced at the Vanderbilt University Medical Center Sequencing Core Facility and the University of Arizona Genomic Analysis and Technology Core Facility, with either Invitrogen vector primer IVT7 or M13R. Resulting sequences were then compared against known Bartonella sequences in GenBank, using default parameters in BLAST.
Phylogenetic methods.
We determined the phylogenetic affinities of the gltA amplicons by first constructing a backbone phylogeny of 18 Bartonella species, isolated from a wide range of mammalian hosts from each of the five recognized host clusters (Table 1). Initially, species were selected by the availability of sequences in public databases from seven housekeeping genes commonly used in Bartonella species delineation (16S, ITS, ftsZ, gltA, groEL, ribC, and rpoB; not all gene sequences were available for all species). However, 16S and ITS were not used because of strong phylogenetic incongruence and alignment uncertainty in these loci. The remaining genes were first aligned using a partial order alignment algorithm (implemented in the software package POA v.2, using default parameters [37]) and then checked by eye for obvious discrepancies. Individual alignments were then concatenated, yielding a global ca. 4.6-kb alignment.
TABLE 1.
Loci and GenBank accession numbers used in the present study to reconstruct Bartonella phylogenetic relationships and to identify the species relationships of cloned gltA amplicons from Polygenis fleas
Maximum parsimony trees were constructed in PAUP 4.0b10 (53), using simple sequence addition, the TBR swapping algorithm, and 10 random addition replicates for each search iteration. Parsimony trees were compared to those generated by a partitioned Bayesian analysis in the software package MrBayes v.3.1.2 (50), under models and parameters separately estimated via Modeltest v.3.6 (45), each conditioned on the same starting tree estimated by maximum likelihood on the entire data set using a general time-reversible model of evolution (6). Priors were not changed from default values. We ran four simultaneous Metropolis-coupled Monte Carlo Markov chains for 1,000,000 generations, with a heating parameter of 0.1. We sampled every 100 generations and calculated a consensus topology after a “burn-in” of 2,500 trees. The consensus tree was then used as a backbone constraint, and cloned gltA amplicons from the Polygenis samples of Bartonella were grafted onto the tree using a simple distance-based neighbor-joining algorithm. For most positive Polygenis samples, this involved replicates of >5 Bartonella clones per individual flea. A newly described species, Bartonella rochalimae, which is closely related to B. clarridgeiae (18), was also grafted onto the tree using a gltA sequence from GenBank (accession no. DQ683195).
pap31 sample size was smaller, and there are generally fewer data available on the gene in Bartonella. Positive sequences were simply aligned with known Bartonella orthologs from GenBank, and both Bayesian and maximum likelihood trees were constructed using the Bayesian methodology described above and for the maximum likelihood tree using a general time-reversible model of nucleotide evolution with parameters estimated from the data. The maximum likelihood analysis was performed with the software program Garli v0.95 (www.bio.utexas.edu/faculty/antisense/garli/Garli.html) (57).
Nucleotide sequence accession numbers.
The gltA and pap31 sequences have been deposited in GenBank under the accession numbers EF616644 to EF616819 and EF625688 to EF625816, respectively.
RESULTS
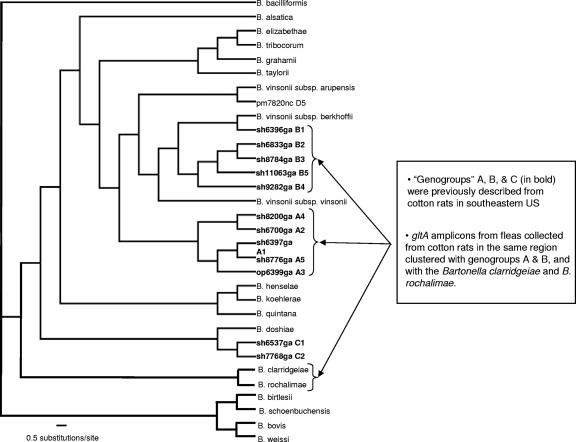
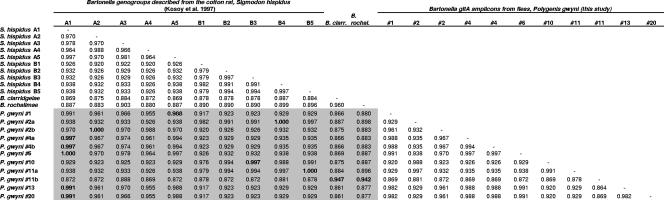
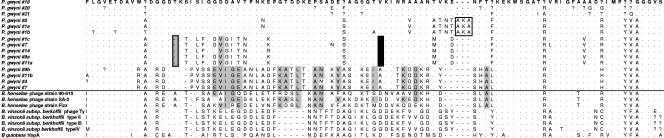
Eight S. hispidus and two N. floridana rats were trapped from two sites, from which 21 P. gwyni fleas were collected. Either gltA or pap31 amplicons of the expected size were detected in 20 of 21 fleas, and replicate sequences were obtained from most samples, such that any Taq error or PCR recombination could be identified and not included in the diversity estimates (Table 2). In 17 fleas, we sequenced multiple and divergent gltA or pap31 clones, permitting us to survey the frequency of single or multiple infections. Cloning efficiency varied between individual fleas, resulting in an unequal number of sequences per gene per flea (Table 2). There was some, but not perfect, overlap between gltA and pap31 as positive evidence for mixed infections. Using gltA only, mixed Bartonella infections were detected in 12 of 16 fleas positive for Bartonella and for which five or more sequence replicates were obtained (Table 2). Most gltA amplicons exhibited greater than 94% sequence similarity to undescribed Bartonella vinsonii-like genogroups previously cultured from S. hispidus in the southeastern United States (Fig. 1 and 2) (33). However, in six fleas collected from five different S. hispidus isolates, gltA amplicons were detected with closest similarity to B. clarridgeiae, a species nominally associated with felines and ruminants (44) but which may be closely related to species with broader host ranges (18, 39). BLAST searches with the pap31 sequences resulted in highest sequence similarity scores with either B. quintana or B. henselae, although neither of these species was greater than 94% similar to any of the pap31 sequences (Fig. 3 and 4), reflecting the still-limited survey of pap31 diversity in Bartonella available in GenBank. However, the pap31 phylogeny revealed three distinct clades of P. gwyni-associated Bartonella, perhaps mirroring the divergence between strains detected by gltA. One clade was characterized by a 1-bp deletion near the boundary between a putative conserved transmembrane domain and an extracellular loop sequence (Fig. 4) (42), producing a UAA stop codon downstream and thus presumably a truncated protein. This deletion was perfectly matched by an alanine-to-valine replacement downstream in a putative inner membrane loop sequence.
TABLE 2.
Identification of Bartonella spp. isolates cloned from each flea, based on reconstruction of gltA or pap31 phylogenies using GenBank sequences and those derived from the present studya
| P. gwynii sample no. | Host | Mammal ID | Site | Distinct gltA genogroupb
|
nd | gltA GenBank accession no(s) | Distinct pap31 cladec
|
nd | pap31 GenBank accession no(s) | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A1/A5 | B2/B3 | B4 | B. clarridgeiae | I | II | III | ||||||||
| 1 | S. hispidus | AEA 10-17 | A | + | + | + | 12 | EF616644/655 | + | + | 8 | EF625688/695 | ||
| 2 | S. hispidus | AEA 10-17 | A | + | + | 5 | EF616656/660 | - | ||||||
| 3 | S. hispidus | AEA 11-20 | A | - | - | |||||||||
| 4 | S. hispidus | AEA 11-20 | A | + | + | 8 | EF616661/668 | + | 3 | EF625696/698 | ||||
| 5 | N. floridana | AEA 01-13 | B | + | + | 12 | EF616669/680 | + | 1 | EF625699 | ||||
| 6 | N. floridana | AEA 01-14 | B | + | 11 | EF616681/691 | - | |||||||
| 7 | S. hispidus | LAD-3331 | B | + | + | 2 | EF616692/693 | + | + | 9 | EF625700/708 | |||
| 8 | S. hispidus | LAD-3331 | B | + | 10 | EF616694/703 | + | 2 | EF625709/710 | |||||
| 9 | S. hispidus | LAD-3331 | B | - | + | + | 6 | EF625711/716 | ||||||
| 10 | S. hispidus | LAD-3332 | B | + | + | 7 | EF616704/710 | + | + | 8 | EF625717/724 | |||
| 11 | S. hispidus | LAD-3332 | B | + | + | + | 7 | EF616711/717 | + | + | + | 5 | EF625725/729 | |
| 12 | S. hispidus | LAD-3333 | B | + | + | 2 | EF616718/719 | + | + | 7 | EF625730/736 | |||
| 13 | S. hispidus | LAD-3333 | B | + | 7 | EF616720/726 | + | 1 | EF625737 | |||||
| 14 | S. hispidus | LAD-3333 | B | + | 8 | EF616727/734 | + | + | 3 | EF625738/740 | ||||
| 15 | S. hispidus | LAD-3333 | B | + | + | 9 | EF616735/742 | + | + | 2 | EF625741/742 | |||
| 16 | S. hispidus | LAD-3334 | B | + | 1 | EF616643 | + | + | 7 | EF625743/749 | ||||
| 17 | S. hispidus | LAD-3334 | B | + | + | 7 | EF616744/750 | + | + | 10 | EF625750/759 | |||
| 18 | S. hispidus | LAD-3334 | B | + | + | + | 15 | EF616751/765 | + | + | + | 15 | EF625760/774 | |
| 19 | S. hispidus | LAD-3335 | B | + | + | 6 | EF616766/771 | + | 11 | EF625775/785 | ||||
| 20 | S. hispidus | LAD-3335 | B | + | + | + | 29 | EF616772/800 | + | + | + | 16 | EF625786/801 | |
| 21 | S. hispidus | LAD-3335 | B | + | + | 19 | EF616801/819 | + | + | + | 15 | EF625802/816 | ||
The two collection sites were Candler County, GA (A) and Bulloch County, GA (B). The gltA columns represent the full diversity of positive matches based on phylogenetic reconstruction. Most matched previously undescribed “genogroups” from Sigmodon hispidus, as shown in Fig. 1 and Fig. 3. The plus signs indicate a positive match. The pap31 clades are provisional designations based on the topology depicted in Fig. 3.
See Fig. 1.
See Fig. 3.
Number of clones sequenced.
FIG. 1.
Bayesian phylogeny of the genus Bartonella, including many of the described species. The tree is rooted with B. bacilliformis and is based on partial sequences from five concatenated loci, with the exception of those shown in bold (see Table 1 for GenBank accession numbers). The bold taxa represent type isolates of the genogroups (designated A thru D) discovered in previous surveys of cotton rats in the southeastern United States (32, 33). Only gltA sequences are available for these. All nonterminal resolved nodes had clade credibility values of >98, based on the Bayesian analysis. The overall topology was supported by parsimony analysis. Arrows indicate the phylogenetic placement of the different P. gwyni-derived isolates on the constrained Bartonella phylogeny, based on neighbor-joining placement of the amplicons on the tree. With the exception of the isolates similar to B. clarridgeiae and B. rochalimae, most were >99% similar to the designated A or B genogroup. Most amplicons were confirmed by redundant sequencing of multiple cloned products.
FIG. 2.
Matrix of genetic similarity (fraction of identical sites) in a 337-bp fragment of gltA cloned from Polygenis gwyni fleas collected from cotton rats (Sigmodon hispidus). Only representative flea-Bartonella samples are shown. Many fleas contained mixed infections, and examples are shown (numbers 2, 4, and 11). The grey shading highlights the values that compare the P. gwyni strains to the corresponding strains A1 through A5 and B1 through B5 previously cultured from S. hispidus (33). The bold values represent the highest similarity values for each flea-associated Bartonella sample to the various A or B genogroups described from cotton rats. B. clarridgeiae and the newly described B. rochalimae (18) are flagellated species distantly related to Bartonella species described from cotton rats but nevertheless are genetically similar to amplicons from the fleas of cotton rats.
FIG. 3.
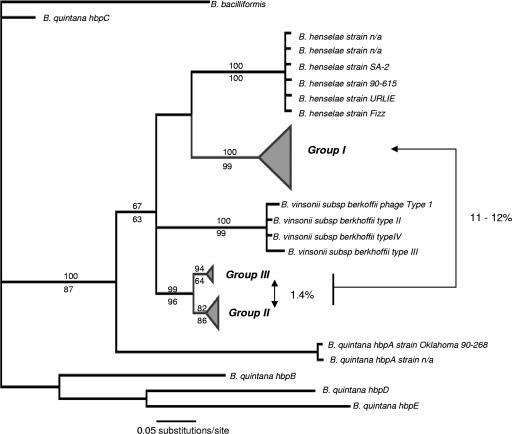
Consensus tree of Bartonella taxa based on partial pap31 sequences. Numbers above interior branches represent clade credibility values from the Bayesian analysis (above) and 100 maximum likelihood bootstrap replicates (below). Most amplicons were confirmed by redundant sequencing of multiple cloned products. Three distinct genotypic groups are evident from the fleas of cotton rats, as shown. The genetic distance between groups II and III, based on 152 bp of alignable transmembrane domain sequences, was approximately 1.4% (uncorrected p distance). Group I differed from both by approximately 11 to 12%. The Bartonella isolates in these groups have no clear identity based on BLAST searches of pap31 sequences. Highest BLAST scores were returned for B. henselae or B. quintana, but this probably reflects the limited taxonomic sampling of pap31 across the genus.
FIG. 4.
Amino acid fragments of pap31 homologs from various taxa and a subset amplified from representative flea samples in the present study. The fragment corresponds to an outer membrane loop sequence between conserved transmembrane domains 3 and 4, as described in reference 42. These loops may be composed of nearly random host chromosomal sequences (56), as evident in the lack of conservation between the related B. henselae and B. quintana. Seven of the nine P. gwyni isolates exhibit strong conservation, likely indicating a recent common ancestor. Some samples align more closely to B. henselae strains, as indicated by gray shading. In one group (clade II from Fig. 3), a 1-bp deletion (grey hatched box) causes a UAA stop codon downstream. A gap has been inserted to maintain the alignment in these samples. The black box highlights an insertion of three residues. Dots indicate matches with the topmost sequence, dashes indicate gaps, and question marks indicate uncertainties due to unresolvable ambiguities in the nucleotide sequences. The black line separates the pap31 amplicons from the present study and those from known species. Dissimilar amplicons were cloned from the same flea samples, as illustrated by P. gwyni numbers 1, 9, and 11.
DISCUSSION
We surveyed the prevalence of Bartonella in a population of rodent fleas, collected from a general locale in which small mammals had been previously intensively surveyed (31, 32, 33). Because we surveyed in a manner that discriminated between single and mixed infections in fleas, we also estimated the fraction of fleas harboring more than one Bartonella isolate and the phylogenetic affinities of coinfecting isolates. We found four noteworthy results.
First, the prevalence of Bartonella was surprisingly high, exceeding characteristic records from various putative arthropod vectors (35, 49, 54). Estimating prevalence requires population-level sampling, and only in recent years have such surveys of Bartonella in presumed vectors begun to emerge. Cat fleas (Ctenocephalides felis) are important agents of zoonotic Bartonella transmission and have been examined in a number of studies sufficient to yield population-level data (41, 49). Estimates of cat-associated Bartonella prevalence (e.g., B. henselae, B. quintana, B. koehlerae, and B. clarridgeiae) have ranged from 20 to 30%, although some C. felis populations may exhibit higher rates (35). Less is known about other flea or arthropod vectors from natural populations of mammals. Studies reporting nonnegligible rates of infection in fleas from various small mammals typically have ranged from 10 to 40% (41, 54). Not surprisingly, there are still few studies that report simultaneous estimates of prevalence in vectors and their mammalian hosts (54).
However, we collected fleas from S. hispidus in an area that, because of extensive prior work (31, 32, 33), corresponds to an intensively scrutinized regional population (the coastal plain and piedmont of Georgia). In one study, Kosoy et al. (32) found rates of Bartonella infection in S. hispidus in central and southern Georgia approaching 80%. Thus, the degree of Bartonella infection in the P. gwyni population we surveyed is consistent with more extensive surveys of S. hispidus and lends confidence that these small sample estimates are representative.
Second, we found substantial rates of mixed Bartonella infections. More than half of the fleas we surveyed were infected by more than one Bartonella gltA genotype (Table 2). If the pap31 screens are included, the rate is even higher. Kosoy et al. (33) originally described four broad genotypic clusters associated with various small rodents from the southeastern United States, designated A through D. Type sequences originally used to define groups A and B together form a diverse but monophyletic group of B. vinsonii-like isolates from S. hispidus, as originally indicated in the neighbor-joining distance gltA tree of Kosoy et al. (33). We detected isolates similar to A and B in the surveyed fleas and, unsurprisingly, did not detect the Peromyscus-associated D group. Surprisingly, we did not detect genogroup C, previously cultured from regional samples of S. hispidus (31, 32). Cluster A is, by far, the most common Bartonella genogroup isolated from cotton rats in the region (31, 32). However, C is more prevalent than B (31, 32), a pattern opposite of what we found in P. gwyni from cotton rats (Table 2; Fig. 1). This pattern may simply be an artifact of small sample sizes and may not hold up to more-extensive surveys. However, one possibility is that the different P. gwyni/S. hispidus isolates exhibit either unequal resident times in the vectors and hosts and/or transmission biases, potentially presenting an opportunity to uncover differential adaptation and specificity in Bartonella (M. Kosoy, personal communication).
Third, we successfully amplified a fragment similar to the heme-binding pap31 in fleas evidently infected with B. vinsonii-like isolates (as determined by gltA). This is notable, because in both B. quintana and B. henselae, pap31 is generally known to be phage-borne and orthologous to a large family of heparin-binding protein-coding genes (hbp) critical to heme acquisition, cellular adhesion, and possibly pathogenesis (8, 11, 56). Recent work has described a pap31 homolog from bacteriophages in B. vinsonii subsp. berkhoffii (40), a species that was previously thought to lack bacteriophages and hbpA protein homologs (8). Assuming the gltA results are a reliable guide, we found pap31-like sequences in fleas infected by Kosoy et al. genogroups A and B; pap31 amplicons were cloned in fleas apparently lacking coinfections and harboring either the A or B gltA genogroup alone (Table 2). Although there is not yet sufficient coverage of the genus with pap31 to identify the isolates we detected, three distinct genogroups are evident (Fig. 3). Possibly, the two derived genogroups within the clade correspond to Kosoy et al.'s (33) genogroups A and B.
In B. quintana, pap31 is a member of a five-gene family, composed of three tandemly arrayed paralogs and two other homologs (42). A possible complication is the uncertain copy number of pap31 homologs across the genus. However, the clade that includes the P. gwyni samples is rooted by hbpA from B. quintana, to the exclusion of other members of the gene family, and includes orthologous sequences from B. henselae and B. vinsonii subsp. berkhoffi (Fig. 3). Moreover, the pap31 transmembrane protein includes outer membrane loops with the potential to incorporate nearly random in-frame chromosomal sequences. In B. quintana, the five homologs are difficult to align at these sites (42) (data not shown). With the exception of some isolates exhibiting distinct similarity to B. henselae (Fig. 4), the loop sequences from P. gwyni isolates exhibit very little amino acid polymorphism between the conserved transmembrane domains. It is thus likely that the pap31 topology reflects orthologous sequence variation in the Bartonella isolates we surveyed. Because of the sampling design and the high rate of coinfection, it is not possible to determine the significance of the truncated hbpA pseudogene. However, possibilities include that some isolates harbor an antigenic variant of the full hbpA protein, similar to the msp2 locus in Anaplasma sp. (3, 7), or that the pap31 pseudogene is a loss-of-function mutant derived from a cryptic strain that has undergone a change in lifestyle (23, 43).
In this vein, the fourth and perhaps most surprising result was the presence in two fleas of an isolate sharing >94% gltA similarity to B. clarridgeiae and the newly described species B. rochalimae (the next closest relative is Bartonella bovis, at >87% sequence similarity) (Fig. 1). B. clarridgeiae itself has not been described from rodents; rather, felids or canids are the primary reservoirs (49). Species near B. clarridgeiae have been reported in various mammalian hosts, however, and recently, a B. clarridgeiae-like isolate was identified in rat fleas from Egypt (39). Among the highest BLAST scores for the B. clarridgeiae-like isolates in Polygenis were uncultured species from rodents and other small mammals (17, 25, 46). Both fleas were coinfected with Kosoy genogroup A or B. In the case of one flea, all three principle gltA variants were detected. The significance of a B. clarridgeiae species in Sigmodon is unknown; because of the size of the survey and the absence of simultaneous information on the competence of Polygenis as a B. clarridgeiae vector and B. clarridgeiae bacteremia in cotton rats, the biological significance is difficult to judge. However, like B. bacilliformis, the etiological agent of bartonellosis (Carrion's disease) in humans, and B. bovis, B. clarridgeiae is one of the few flagellated bartonellae (51) and has long been a problematic species because of its uncertain phylogenetic placement and the odd host range that it shares with B. bovis (44). It may not be a coincidence that an isolate resembling these hyper-generalist species has been discovered in Polygenis. Efforts to understand the molecular basis of variation in host specificity in the genus ( 2) would benefit from closer examination of B. clarridgeiae and its relatives (18).
Two opposing ecological and evolutionary processes seem to be at work in Bartonella. The cryptic diversity in the vectors of Bartonella, and the absence of strains common in mammalian hosts, may reflect an evolutionary trend towards differential adaptation to host-specific niches, in either vectors or reservoirs. If so, it seems Bartonella possesses a tendency towards fine-scale adaptation, ecological specialization, and divergence between essentially syntopic populations, despite mixed infections, close physical proximity, and generalist lifestyles. The mechanisms by which Bartonella genomes are protected during the process of specialization to host-associated niches, while maintaining broad host affiliations and thus mixed infections (29), are presently unknown.
Acknowledgments
We thank Michael Kosoy for helpful discussion, as well as three anonymous reviewers for helpful comments on early drafts.
This work was supported by NSF grant IOB-0429400 to P.A.
Footnotes
Published ahead of print on 10 August 2007.
REFERENCES
- 1.Alexander, B. 1995. A review of bartonellosis in Ecuador and Colombia. Am. J. Trop. Med. Hyg. 52:354-359. [DOI] [PubMed] [Google Scholar]
- 2.Alsmark, C. M., A. C. Frank, E. O. Karlberg, B. A. Legault, D. H. Ardell, B. Canback, A. S. Eriksson, A. K. Naslund, S. A. Handley, M. Huvet, B. La Scola, M. Holmberg, and S. G. E. Andersson. 2004. The louse-borne human pathogen Bartonella quintana is a genomic derivative of the zoonotic agent Bartonella henselae. Proc. Acad. Natl. Sci. USA 101:9716-9721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barbet, A. F., P. F. M. Meeus, M. Belanger, M. V. Bowie, J. Yi, A. M. Lundgren, A. R. Alleman, S. J. Wong, F. K. Chu, U. G. Munderloh, and S. D. Jauron. 2003. Expression of multiple outer membrane protein sequence variants from a single genomic locus of Anaplasma phagocytophilum. Infect. Immun. 71:1706-1718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boucher, Y., C. J. Douady, R. T. Papke, D. A. Walsh, M. E. R. Boudreau, C. L. Nesbo, R. J. Case, and W. F. Doolittle. 2003. Lateral gene transfer and the origins of prokaryotic groups. Annu. Rev. Genet. 37:283-328. [DOI] [PubMed] [Google Scholar]
- 5.Boulouis, H. J., C. C. Chang, J. B. Henn, R. W. Kasten, and B. B. Chomel. 2005. Factors associated with the rapid emergence of zoonotic Bartonella infections. Vet. Res. 36:383-410. [DOI] [PubMed] [Google Scholar]
- 6.Brandley, M. C., A. Schmitz, and T. W. Reeder. 2005. Partitioned Bayesian analyses, partition choice, and the phylogenetic relationships of scincid lizards. Syst. Biol. 54:373-390. [DOI] [PubMed] [Google Scholar]
- 7.Brayton, K. A., D. P. Knowles, T. C. McGuire, and G. H. Palmer. 2001. Efficient use of a small genome to generate antigenic diversity in tick-borne ehrlichial pathogens. Proc. Acad. Natl. Sci. USA 98:4130-4135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carroll, J. A., S. A. Coleman, L. S. Smitherman, and M. F. Minnick. 2000. Hemin-binding surface protein from Bartonella quintana. Infect. Immun. 68:6750-6757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chomel, B. B., R. W. Kasten, K. Floyd Hawkins, B. H. Chi, K. Yamamoto, J. Roberts Wilson, A. N. Gurfield, R. C. Abbott, N. C. Pedersen, and J. E. Koehler. 1996. Experimental transmission of Bartonella henselae by the cat flea. J. Clin. Microbiol. 34:1952-1956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cox, F. E. G. 2001. Concomitant infections. Parasitology 122:S1. [DOI] [PubMed] [Google Scholar]
- 11.Dabo, S. M., A. W. Confer, B. E. Anderson, and S. Gupta. 2006. Bartonella henselae Pap31, an extracellular matrix adhesin, binds the fibronectin repeat III13 module. Infect. Immun. 74:2513-2521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dehio, C. 2005. Bartonella-host-cell interactions and vascular tumour formation. Nat. Rev. Microbiol. 3:621-631. [DOI] [PubMed] [Google Scholar]
- 13.Dehio, C. 2001. Bartonella interactions with endothelial cells and erythrocytes. Trends Microbiol. 9:279-285. [DOI] [PubMed] [Google Scholar]
- 14.Dehio, C. 2004. Molecular and cellular basis of Bartonella pathogenesis. Annu. Rev. Microbiol. 58:365-390. [DOI] [PubMed] [Google Scholar]
- 15.de Roode, J. C., M. E. H. Helinski, M. A. Anwar, and A. F. Read. 2005. Dynamics of multiple infection and within-host competition in genetically diverse malaria infections. Am. Nat. 166:531-542. [DOI] [PubMed] [Google Scholar]
- 16.Durden, L. A., C. W. Banks, K. L. Clark, B. V. Belbey, and J. H. Oliver, Jr. 1997. Ectoparasite fauna of the eastern woodrat, Neotoma floridana: composition, origin, and comparison with ectoparasite faunas of western woodrat species. J. Parasitol. 83:374-381. [PubMed] [Google Scholar]
- 17.Ehrenborg, C., S. Handley, B. A. Ellis, J. N. Mills, and M. Holmberg. 2003. Bartonella grahamii infecting rodents display high genetic diversity over short geographic distances. Ann. N. Y. Acad. Sci. 990:233-235. [DOI] [PubMed] [Google Scholar]
- 18.Eremeeva, M. E., H. L. Gerns, S. L. Lydy, J. S. Goo, E. T. Ryan, S. S. Mathew, M. J. Ferraro, J. M. Holden, W. L. Nicholson, G. A. Dasch, and J. E. Koehler. 2007. Bacteremia, fever, and sphenomegaly caused by a newly recognized Bartonella species. N. Engl. J. Med. 356:2381-2387. [DOI] [PubMed] [Google Scholar]
- 19.Finkelstein, J. L., T. P. Brown, K. L. O'Reilly, J. Wedincamp, and L. D. Foil. 2002. Studies on the growth of Bartonella henselae in the cat flea (Siphonaptera: Pulicidae). J. Med. Entomol. 39:915-919. [DOI] [PubMed] [Google Scholar]
- 20.Foil, L., E. Andress, R. L. Freeland, A. F. Roy, R. Rutledge, P. C. Triche, and K. L. O'Reilly. 1998. Experimental infection of domestic cats with Bartonella henselae by inoculation of Ctenocephalides felis (Siphonaptera: Pulicidae) feces. J. Med. Entomol. 35:625-628. [DOI] [PubMed] [Google Scholar]
- 21.Fournier, P. E., J. B. Ndihokubwayo, J. Guidran, P. J. Kelly, and D. Raoult. 2002. Human pathogens in body and head lice. Emerg. Infect. Dis. 8:1515-1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gurfield, A. N., H. J. Boulouis, B. B. Chomel, R. Heller, R. W. Kasten, K. Yamamoto, and Y. Piemont. 1997. Coinfection with Bartonella clarridgeiae and Bartonella henselae and with different Bartonella henselae strains in domestic cats. J. Clin. Microbiol. 35:2120-2123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Halos, L., T. Jamal, R. Maillard, B. Girard, J. Guillot, B. Chomel, M. Vayssier-Taussat, and H. J. Boulouis. 2004. Role of Hippoboscidae flies as potential vectors of Bartonella spp. infecting wild and domestic ruminants. Appl. Environ. Microbiol. 70:6302-6305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Harvell, C. D., C. E. Mitchell, J. R. Ward, S. Altizer, A. P. Dobson, R. S. Ostfeld, and M. D. Samuel. 2002. Ecology: climate warming and disease risks for terrestrial and marine biota. Science 296:2158-2162. [DOI] [PubMed] [Google Scholar]
- 25.Hinnebusch, B. J., M. L. Rosso, T. G. Schwan, and E. Carniel. 2002. High-frequency conjugative transfer of antibiotic resistance genes to Yersinia pestis in the flea midgut. Mol. Microbiol. 46:349-354. [DOI] [PubMed] [Google Scholar]
- 26.Holmberg, M., J. N. Mills, S. McGill, G. Benjamin, and B. A. Ellis. 2003. Bartonella infection in sylvatic small mammals of central Sweden. Epidemiol. Infect. 130:149-157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Iredell, J., D. Blanckenberg, M. Arvand, S. Grauling, E. J. Feil, and R. J. Birtles. 2003. Characterization of the natural population of Bartonella henselae by multilocus sequence typing. J. Clin. Microbiol. 41:5071-5079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jacomo, V., P. J. Kelly, and D. Raoult. 2002. Natural history of Bartonella infections (an exception to Koch's postulate). Clin. Diagn. Lab. Immunol. 9:8-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kang, J., and M. J. Blaser. 2006. Bacterial populations as perfect gases: genomic integrity and diversification tensions in Helicobacter pylori. Nat. Rev. Microbiol. 4:826-836. [DOI] [PubMed] [Google Scholar]
- 30.Karem, K. L. 2000. Immune aspects of Bartonella. Crit. Rev. Microbiol. 26:133-145. [DOI] [PubMed] [Google Scholar]
- 31.Kosoy, M., E. Mandel, D. Green, E. Marston, and J. E. Childs. 2004. Prospective studies of Bartonella of rodents. Part I. Demographic and temporal patterns in population dynamics. Vector-Borne Zoonot. Dis. 4:285-295. [DOI] [PubMed] [Google Scholar]
- 32.Kosoy, M., E. Mandel, D. Green, E. Marston, D. C. Jones, and J. E. Childs. 2004. Prospective studies of Bartonella of rodents. Part II. Diverse infections in a single rodent community. Vector-Borne Zoonot. Dis. 4:296-305. [DOI] [PubMed] [Google Scholar]
- 33.Kosoy, M. Y., R. L. Regnery, T. Tzianabos, E. L. Marston, D. C. Jones, D. Green, G. O. Maupin, J. G. Olson, and J. E. Childs. 1997. Distribution, diversity, and host specificity of Bartonella in rodents from the southeastern United States. Am. J. Trop. Med. Hyg. 57:578-588. [DOI] [PubMed] [Google Scholar]
- 34.Kurtenbach, K., K. Hanincova, J. I. Tsao, G. Margos, D. Fish, and N. H. Ogden. 2006. Fundamental processes in the evolutionary ecology of Lyme borreliosis. Nat. Rev. Microbiol. 4:660-669. [DOI] [PubMed] [Google Scholar]
- 35.Lappin, M. R., B. Griffin, J. Brunt, A. Riley, D. Burney, J. Hawley, M. M. Brewer, and W. A. Jensen. 2006. Prevalence of Bartonella species, Haemoplasma species, Ehrlichia species, Anaplasma phagocytophilum, and Neorickettsia risticii DNA in the blood of cats and their fleas in the United States. J. Feline Med. Surg. 8:85-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.La Scola, B., Z. Zeaiter, A. Khamis, and D. Raoult. 2003. Gene-sequence-based criteria for species definition in bacteriology: the Bartonella paradigm. Trends Microbiol. 11:318-321. [DOI] [PubMed] [Google Scholar]
- 37.Lee, C., C. Grasso, and M. F. Sharlow. 2002. Multiple sequence alignment using partial order graphs. Bioinformatics 18:452-464. [DOI] [PubMed] [Google Scholar]
- 38.Lewis, R. E., and J. H. Lewis. 1994. Siphonaptera of North America north of Mexico—Vermipsyllidae and Rhopalopsyllidae. J. Med. Entomol. 31:82-98. [DOI] [PubMed] [Google Scholar]
- 39.Loftis, A. D., W. K. Reeves, D. E. Szumlas, M. M. Abbassy, I. M. Helmy, J. R. Mortarity, and G. A. Dasch. 2006. Surveillance of Egyptian fleas for agents of public health significance: Anaplasma, Bartonella, Coxiella, Ehrlichia, Rickettsia, and Yersinia pestis. Am. J. Trop. Med. Hyg. 75:41-48. [PubMed] [Google Scholar]
- 40.Maggi, R. G., and E. B. Breitschwerdt. 2005. Isolation of bacteriophages from Bartonella vinsonii subsp. berkhoffii and the characterization of Pap31 gene sequences from bacterial and phage DNA. J. Mol. Microbiol. Biotechnol. 9:44-51. [DOI] [PubMed] [Google Scholar]
- 41.Marie, J. L., P. E. Fournier, J. M. Rolain, S. Briolant, B. Davoust, and D. Raoult. 2006. Molecular detection of Bartonella quintana, B. elizabethae, B. koehlerae, B. doshiae, B. taylorii, and Rickettsia felis in rodent fleas collected in Kabul, Afghanistan. Am. J. Trop. Med. Hyg. 74:436-439. [PubMed] [Google Scholar]
- 42.Minnick, M. F., K. N. Sappington, L. S. Smitherman, S. G. E. Andersson, O. Karlberg, and J. A. Carroll. 2003. Five-member gene family of Bartonella quintana. Infect. Immun. 71:814-821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Noda, H., U. G. Munderloh, and T. J. Kurtti. 1997. Endosymbionts of ticks and their relationship to Wolbachia spp. and tick-borne pathogens of humans and animals. Appl. Environ. Microbiol. 63:3926-3932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pitulle, C., C. Strehse, J. W. Brown, and E. B. Breitschwerdt. 2002. Investigation of the phylogenetic relationships within the genus Bartonella based on comparative sequence analysis of the rnpB gene, 16S rDNA and 23S rDNA. Int. J. Syst. Evol. Microbiol. 52:2075-2080. [DOI] [PubMed] [Google Scholar]
- 45.Posada, D., and K. A. Crandall. 1998. MODELTEST: testing the model of DNA substitution. Bioinformatics 14:817-818. [DOI] [PubMed] [Google Scholar]
- 46.Pretorius, A. M., L. Beati, and R. J. Birtles. 2004. Diversity of bartonellae associated with small mammals inhabiting Free State province, South Africa. Int. J. Syst. Evol. Microbiol. 54:1959-1967. [DOI] [PubMed] [Google Scholar]
- 47.Raberg, L., J. C. de Roode, A. S. Bell, P. Stamou, D. Gray, and A. F. Read. 2006. The role of immune-mediated apparent competition in genetically diverse malaria infections. Am. Nat. 168:41-53. [DOI] [PubMed] [Google Scholar]
- 48.Read, A. F., and L. H. Taylor. 2001. The ecology of genetically diverse infections. Science 292:1099-1102. [DOI] [PubMed] [Google Scholar]
- 49.Rolain, J. M., M. Franc, B. Davoust, and D. Raoult. 2003. Molecular detection of Bartonella quintana, B. koehlerae, B. henselae, B. clarridgeiae, Rickettsia felis, and Wolbachia pipientis in cat fleas, France. Emerg. Infect. Dis. 9:338-342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ronquist, F., and J. P. Huelsenbeck. 2003. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19:1572-1574. [DOI] [PubMed] [Google Scholar]
- 51.Sander, A., A. Zagrosek, W. Bredt, E. Schiltz, Y. Piemont, C. Lanz, and C. Dehio. 2000. Characterization of Bartonella clarridgeiae flagellin (FlaA) and detection of antiflagellin antibodies in patients with lymphadenopathy. J. Clin. Microbiol. 38:2943-2948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Smit, F. G. A. M. 1987. An illustrated catalogue of the Rothschild collection of fleas (Siphonaptera) in the British Museum (Natural History) with keys and short descriptions for the identification of families, genera, species and subspecies of the order. Oxford University Press and The British Museum (Natural History), Oxford, England.
- 53.Swofford, D. L. 1998. PAUP*: phylogenetic analysis using parsimony (*and other methods), version 4.0b10. Sinauer Associates, Sunderland, MA.
- 54.Telfer, S., K. J. Bown, R. Sekules, I. Begon, T. Hayden, and R. Birtles. 2005. Disruption of a host-parasite system following the introduction of an exotic host species. Parasitology 130:661-668. [DOI] [PubMed] [Google Scholar]
- 55.Thrall, P. H., M. E. Hochberg, J. J. Burdon, and J. D. Bever. 2006. Coevolution of symbiotic mutualists and parasites in a community context. Trends Ecol. Evol. 22:120-126. [DOI] [PubMed] [Google Scholar]
- 56.Zimmermann, R., V. A. J. Kempf, E. Schiltz, K. Oberle, and A. Sander. 2003. Hemin binding, functional expression, and complementation analysis of Pap31 from Bartonella henselae. J. Bacteriol. 185:1739-1744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zwickl, D. J. 2006. Genetic algorithm approaches for the phylogenetic analysis of large biological sequence datasets under the maximum likelihood criterion. Ph.D. dissertation, The University of Texas at Austin.