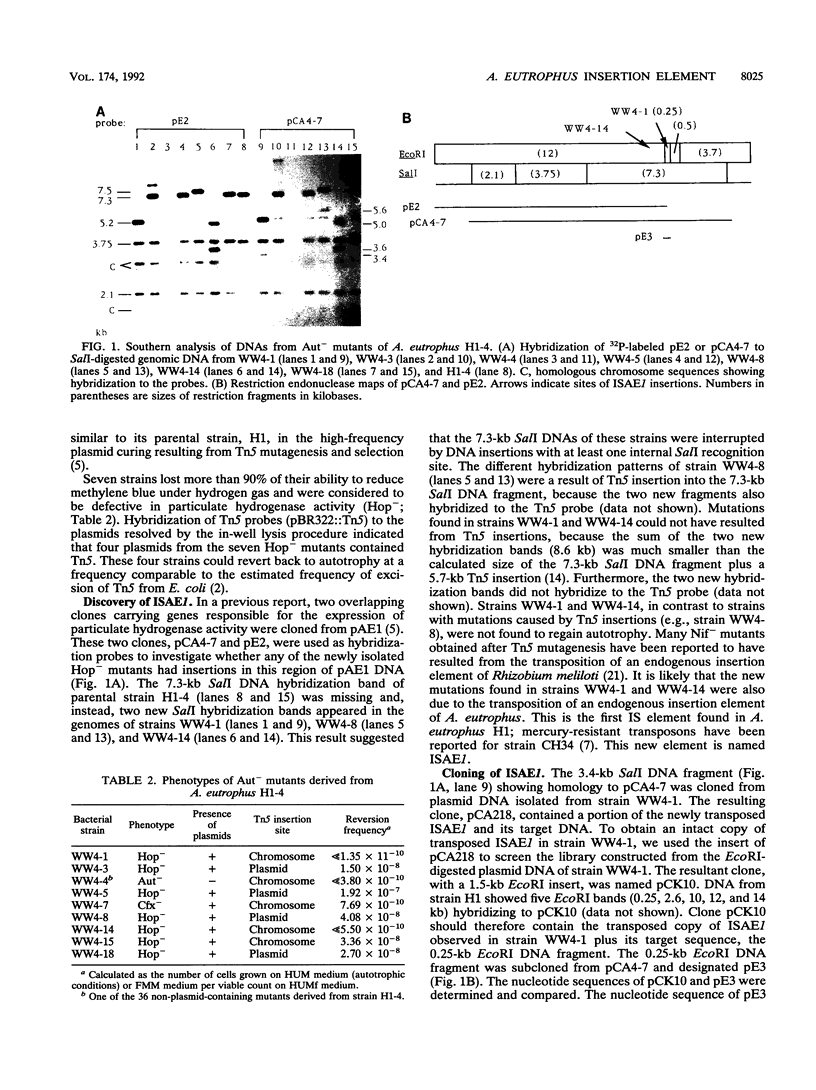
Abstract
An insertion element, ISAE1, was discovered during the molecular analysis of mutants defective in the autotrophic growth (Aut-) of Alcaligenes eutrophus H1-4, a mitomycin C-generated derivative of strain H1. ISAE1 is 1,313 bp long, has 12-bp nearly perfect inverted terminal repeats, and contains an open reading frame that has a coding capacity of 408 amino acids. Direct repeats of 8 bp were generated by insertion of ISAE1 into chromosomes or plasmids. Most insertion were found in the AT-rich target sites. The distribution of ISAE1 is limited to A. eutrophus H1 (ATCC 17698) and H16 (ATCC 17699). Variants with newly transposed copies of ISAE1 could be isolated at an elevated frequency by changing the growth conditions.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Beringer J. E. R factor transfer in Rhizobium leguminosarum. J Gen Microbiol. 1974 Sep;84(1):188–198. doi: 10.1099/00221287-84-1-188. [DOI] [PubMed] [Google Scholar]
- Cai Y. Characterization of insertion sequence IS892 and related elements from the cyanobacterium Anabaena sp. strain PCC 7120. J Bacteriol. 1991 Sep;173(18):5771–5777. doi: 10.1128/jb.173.18.5771-5777.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cirillo J. D., Barletta R. G., Bloom B. R., Jacobs W. R., Jr A novel transposon trap for mycobacteria: isolation and characterization of IS1096. J Bacteriol. 1991 Dec;173(24):7772–7780. doi: 10.1128/jb.173.24.7772-7780.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eberz G., Eitinger T., Friedrich B. Genetic determinants of a nickel-specific transport system are part of the plasmid-encoded hydrogenase gene cluster in Alcaligenes eutrophus. J Bacteriol. 1989 Mar;171(3):1340–1345. doi: 10.1128/jb.171.3.1340-1345.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckhardt T. A rapid method for the identification of plasmid desoxyribonucleic acid in bacteria. Plasmid. 1978 Sep;1(4):584–588. doi: 10.1016/0147-619x(78)90016-1. [DOI] [PubMed] [Google Scholar]
- Gay P., Le Coq D., Steinmetz M., Berkelman T., Kado C. I. Positive selection procedure for entrapment of insertion sequence elements in gram-negative bacteria. J Bacteriol. 1985 Nov;164(2):918–921. doi: 10.1128/jb.164.2.918-921.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogrefe C., Römermann D., Friedrich B. Alcaligenes eutrophus hydrogenase genes (Hox). J Bacteriol. 1984 Apr;158(1):43–48. doi: 10.1128/jb.158.1.43-48.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorgensen R. A., Rothstein S. J., Reznikoff W. S. A restriction enzyme cleavage map of Tn5 and location of a region encoding neomycin resistance. Mol Gen Genet. 1979;177(1):65–72. doi: 10.1007/BF00267254. [DOI] [PubMed] [Google Scholar]
- Maier R. J., Campbell N. E., Hanus F. J., Simpson F. B., Russell S. A., Evans H. J. Expression of hydrogenase activity in free-living Rhizobium japonicum. Proc Natl Acad Sci U S A. 1978 Jul;75(7):3258–3262. doi: 10.1073/pnas.75.7.3258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mergeay M., Nies D., Schlegel H. G., Gerits J., Charles P., Van Gijsegem F. Alcaligenes eutrophus CH34 is a facultative chemolithotroph with plasmid-bound resistance to heavy metals. J Bacteriol. 1985 Apr;162(1):328–334. doi: 10.1128/jb.162.1.328-334.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulligan M. E., Hawley D. K., Entriken R., McClure W. R. Escherichia coli promoter sequences predict in vitro RNA polymerase selectivity. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 2):789–800. doi: 10.1093/nar/12.1part2.789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson W. R., Lipman D. J. Improved tools for biological sequence comparison. Proc Natl Acad Sci U S A. 1988 Apr;85(8):2444–2448. doi: 10.1073/pnas.85.8.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruvkun G. B., Long S. R., Meade H. M., van den Bos R. C., Ausubel F. M. ISRm1: A Rhizobium meliloti insertion sequence that transposes preferentially into nitrogen fixation genes. J Mol Appl Genet. 1982;1(5):405–418. [PubMed] [Google Scholar]
- Schink B., Schlegel H. G. The membrane-bound hydrogenase of Alcaligenes eutrophus. I. Solubilization, purification, and biochemical properties. Biochim Biophys Acta. 1979 Apr 12;567(2):315–324. doi: 10.1016/0005-2744(79)90117-7. [DOI] [PubMed] [Google Scholar]
- Schneider K., Schlegel H. G. Purification and properties of soluble hydrogenase from Alcaligenes eutrophus H 16. Biochim Biophys Acta. 1976 Nov 8;452(1):66–80. doi: 10.1016/0005-2744(76)90058-9. [DOI] [PubMed] [Google Scholar]
- Uffen R. L., Colbeau A., Richaud P., Vignais P. M. Cloning and sequencing the genes encoding uptake-hydrogenase subunits of Rhodocyclus gelatinosus. Mol Gen Genet. 1990 Mar;221(1):49–58. doi: 10.1007/BF00280367. [DOI] [PubMed] [Google Scholar]
- Yanisch-Perron C., Vieira J., Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33(1):103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]




