Abstract
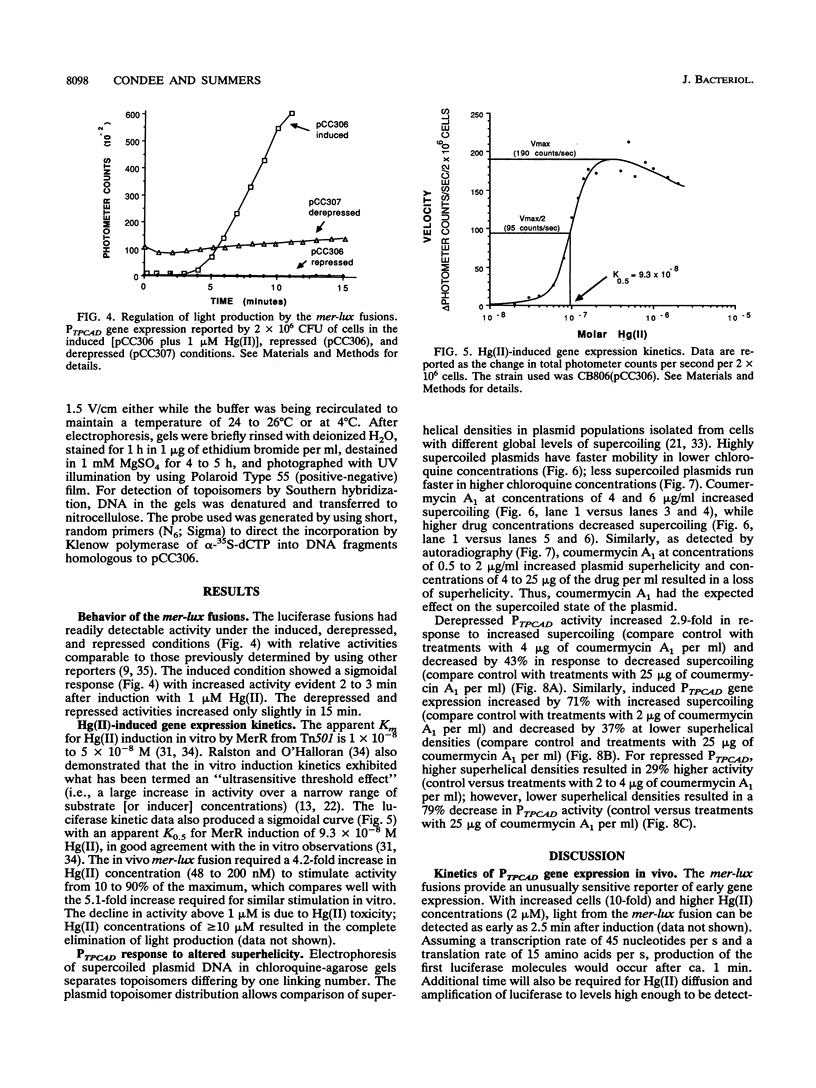
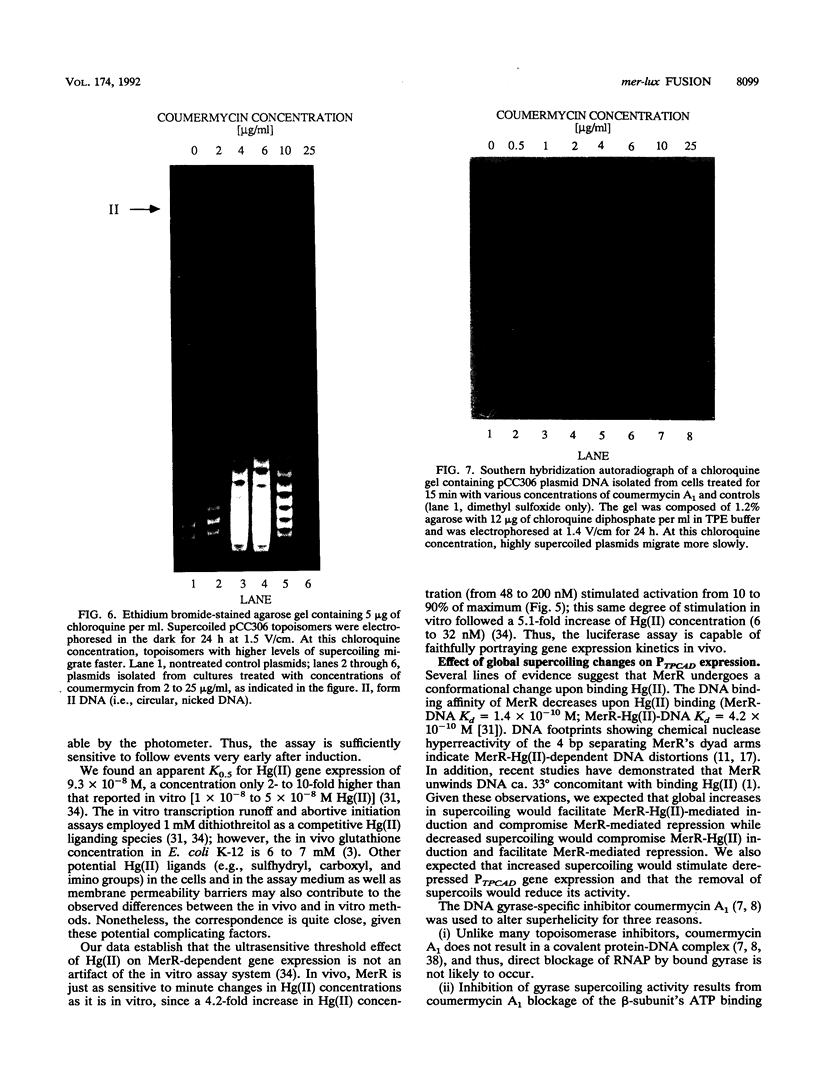
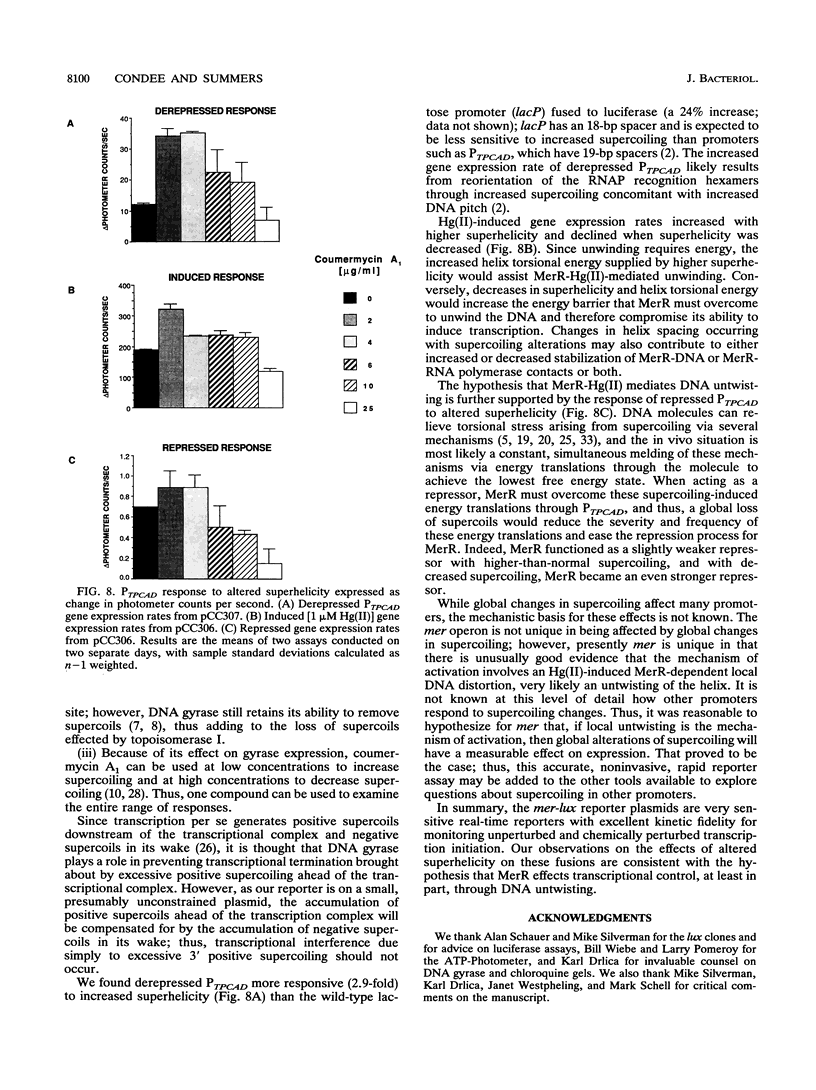
We constructed mercury resistance operon-luciferase (mer-lux) transcriptional fusion plasmids to evaluate in vivo gene expression rates of the mer structural gene promoter (PTPCAD) of transposon Tn21. In vivo gene expression kinetics corresponded well with those previously determined in vitro, yielding an apparent K0.5 for Hg(II)-stimulated induction by MerR of 9.3 x 10(-8) M with the same ultrasensitive threshold effect seen in vitro. We also used the mer-lux fusions to elucidate subtle variations in promoter activity brought about by altered superhelicity. Binding of inducer [Hg(II)] to the transcriptional activator MerR is known to result in DNA distortion and transcriptional activation of the mer operon; it has recently been demonstrated that this distortion is a consequence of MerR-Hg(II)-induced local DNA unwinding to facilitate RNA polymerase open complex formation at PTPCAD. Since negative supercoiling results in DNA unwinding similar to this MerR activation, we hypothesized that a global increase in plasmid supercoiling would facilitate MerR-mediated activation and compromise MerR-mediated repression, while removal of plasmid supercoils would compromise MerR's ability to induce transcription and facilitate its ability to repress transcription. Indeed, we found that increased negative supercoiling results in increased gene expression rates and decreased supercoiling results in reduced gene expression rates for the induced, repressed, and derepressed conditions of PTPCAD. Thus, luciferase transcriptional fusions can detect subtle variations in initial rates of gene expression in a real-time, nondestructive assay.
Full text
PDF

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ansari A. Z., Chael M. L., O'Halloran T. V. Allosteric underwinding of DNA is a critical step in positive control of transcription by Hg-MerR. Nature. 1992 Jan 2;355(6355):87–89. doi: 10.1038/355087a0. [DOI] [PubMed] [Google Scholar]
- Aoyama T., Takanami M. Supercoiling response of E. coli promoters with different spacer lengths. Biochim Biophys Acta. 1988 Mar 31;949(3):311–317. doi: 10.1016/0167-4781(88)90157-1. [DOI] [PubMed] [Google Scholar]
- Apontoweil P., Berends W. Isolation and initial characterization of glutathione-deficient mutants of Escherichia coli K 12. Biochim Biophys Acta. 1975 Jul 14;399(1):10–22. doi: 10.1016/0304-4165(75)90206-8. [DOI] [PubMed] [Google Scholar]
- Birnboim H. C., Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979 Nov 24;7(6):1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boles T. C., White J. H., Cozzarelli N. R. Structure of plectonemically supercoiled DNA. J Mol Biol. 1990 Jun 20;213(4):931–951. doi: 10.1016/S0022-2836(05)80272-4. [DOI] [PubMed] [Google Scholar]
- Depew D. E., Wang J. C. Conformational fluctuations of DNA helix. Proc Natl Acad Sci U S A. 1975 Nov;72(11):4275–4279. doi: 10.1073/pnas.72.11.4275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drlica K., Coughlin S. Inhibitors of DNA gyrase. Pharmacol Ther. 1989;44(1):107–121. doi: 10.1016/0163-7258(89)90093-4. [DOI] [PubMed] [Google Scholar]
- Drlica K., Franco R. J. Inhibitors of DNA topoisomerases. Biochemistry. 1988 Apr 5;27(7):2253–2259. doi: 10.1021/bi00407a001. [DOI] [PubMed] [Google Scholar]
- Foster T. J., Ginnity F. Some mercurial resistance plasmids from different incompatibility groups specify merR regulatory functions that both repress and induce the mer operon of plasmid R100. J Bacteriol. 1985 May;162(2):773–776. doi: 10.1128/jb.162.2.773-776.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franco R. J., Drlica K. Gyrase inhibitors can increase gyrA expression and DNA supercoiling. J Bacteriol. 1989 Dec;171(12):6573–6579. doi: 10.1128/jb.171.12.6573-6579.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frantz B., O'Halloran T. V. DNA distortion accompanies transcriptional activation by the metal-responsive gene-regulatory protein MerR. Biochemistry. 1990 May 22;29(20):4747–4751. doi: 10.1021/bi00472a001. [DOI] [PubMed] [Google Scholar]
- Gambill B. D., Summers A. O. Versatile mercury-resistant cloning and expression vectors. Gene. 1985;39(2-3):293–297. doi: 10.1016/0378-1119(85)90326-9. [DOI] [PubMed] [Google Scholar]
- Goldbeter A., Koshland D. E., Jr An amplified sensitivity arising from covalent modification in biological systems. Proc Natl Acad Sci U S A. 1981 Nov;78(11):6840–6844. doi: 10.1073/pnas.78.11.6840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein E., Drlica K. Regulation of bacterial DNA supercoiling: plasmid linking numbers vary with growth temperature. Proc Natl Acad Sci U S A. 1984 Jul;81(13):4046–4050. doi: 10.1073/pnas.81.13.4046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helmann J. D., Ballard B. T., Walsh C. T. The MerR metalloregulatory protein binds mercuric ion as a tricoordinate, metal-bridged dimer. Science. 1990 Feb 23;247(4945):946–948. doi: 10.1126/science.2305262. [DOI] [PubMed] [Google Scholar]
- Heltzel A., Gambill D., Jackson W. J., Totis P. A., Summers A. O. Overexpression and DNA-binding properties of the mer-encoded regulatory protein from plasmid NR1 (Tn21). J Bacteriol. 1987 Jul;169(7):3379–3384. doi: 10.1128/jb.169.7.3379-3384.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heltzel A., Lee I. W., Totis P. A., Summers A. O. Activator-dependent preinduction binding of sigma-70 RNA polymerase at the metal-regulated mer promoter. Biochemistry. 1990 Oct 16;29(41):9572–9584. doi: 10.1021/bi00493a011. [DOI] [PubMed] [Google Scholar]
- Horwitz M. S., Loeb L. A. An E. coli promoter that regulates transcription by DNA superhelix-induced cruciform extrusion. Science. 1988 Aug 5;241(4866):703–705. doi: 10.1126/science.2456617. [DOI] [PubMed] [Google Scholar]
- Johnston B. H. The S1-sensitive form of d(C-T)n.d(A-G)n: chemical evidence for a three-stranded structure in plasmids. Science. 1988 Sep 30;241(4874):1800–1804. doi: 10.1126/science.2845572. [DOI] [PubMed] [Google Scholar]
- Keller W. Determination of the number of superhelical turns in simian virus 40 DNA by gel electrophoresis. Proc Natl Acad Sci U S A. 1975 Dec;72(12):4876–4880. doi: 10.1073/pnas.72.12.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koshland D. E., Jr, Némethy G., Filmer D. Comparison of experimental binding data and theoretical models in proteins containing subunits. Biochemistry. 1966 Jan;5(1):365–385. doi: 10.1021/bi00865a047. [DOI] [PubMed] [Google Scholar]
- Lathe R., Kieny M. P., Skory S., Lecocq J. P. Linker tailing: unphosphorylated linker oligonucleotides for joining DNA termini. DNA. 1984;3(2):173–182. doi: 10.1089/dna.1984.3.173. [DOI] [PubMed] [Google Scholar]
- Lee I. W., Gambill B. D., Summers A. O. Translation of merD in Tn21. J Bacteriol. 1989 Apr;171(4):2222–2225. doi: 10.1128/jb.171.4.2222-2225.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lilley D. M. In vivo consequences of plasmid topology. Nature. 1981 Jul 23;292(5821):380–382. doi: 10.1038/292380a0. [DOI] [PubMed] [Google Scholar]
- Liu L. F., Wang J. C. Supercoiling of the DNA template during transcription. Proc Natl Acad Sci U S A. 1987 Oct;84(20):7024–7027. doi: 10.1073/pnas.84.20.7024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lund P. A., Ford S. J., Brown N. L. Transcriptional regulation of the mercury-resistance genes of transposon Tn501. J Gen Microbiol. 1986 Feb;132(2):465–480. doi: 10.1099/00221287-132-2-465. [DOI] [PubMed] [Google Scholar]
- Manes S. H., Pruss G. J., Drlica K. Inhibition of RNA synthesis by oxolinic acid is unrelated to average DNA supercoiling. J Bacteriol. 1983 Jul;155(1):420–423. doi: 10.1128/jb.155.1.420-423.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClure W. R. Mechanism and control of transcription initiation in prokaryotes. Annu Rev Biochem. 1985;54:171–204. doi: 10.1146/annurev.bi.54.070185.001131. [DOI] [PubMed] [Google Scholar]
- Mukhopadhyay D., Yu H. R., Nucifora G., Misra T. K. Purification and functional characterization of MerD. A coregulator of the mercury resistance operon in gram-negative bacteria. J Biol Chem. 1991 Oct 5;266(28):18538–18542. [PubMed] [Google Scholar]
- O'Halloran T. V., Frantz B., Shin M. K., Ralston D. M., Wright J. G. The MerR heavy metal receptor mediates positive activation in a topologically novel transcription complex. Cell. 1989 Jan 13;56(1):119–129. doi: 10.1016/0092-8674(89)90990-2. [DOI] [PubMed] [Google Scholar]
- Parkhill J., Brown N. L. Site-specific insertion and deletion mutants in the mer promoter-operator region of Tn501; the nineteen base-pair spacer is essential for normal induction of the promoter by MerR. Nucleic Acids Res. 1990 Sep 11;18(17):5157–5162. doi: 10.1093/nar/18.17.5157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahmouni A. R., Wells R. D. Stabilization of Z DNA in vivo by localized supercoiling. Science. 1989 Oct 20;246(4928):358–363. doi: 10.1126/science.2678475. [DOI] [PubMed] [Google Scholar]
- Ralston D. M., O'Halloran T. V. Ultrasensitivity and heavy-metal selectivity of the allosterically modulated MerR transcription complex. Proc Natl Acad Sci U S A. 1990 May;87(10):3846–3850. doi: 10.1073/pnas.87.10.3846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross W., Park S. J., Summers A. O. Genetic analysis of transcriptional activation and repression in the Tn21 mer operon. J Bacteriol. 1989 Jul;171(7):4009–4018. doi: 10.1128/jb.171.7.4009-4018.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz D. W., Yarus M. A simple and sensitive in vivo luciferase assay for tRNA-mediated nonsense suppression. J Bacteriol. 1990 Feb;172(2):595–602. doi: 10.1128/jb.172.2.595-602.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shewchuk L. M., Verdine G. L., Walsh C. T. Transcriptional switching by the metalloregulatory MerR protein: initial characterization of DNA and mercury (II) binding activities. Biochemistry. 1989 Mar 7;28(5):2331–2339. doi: 10.1021/bi00431a052. [DOI] [PubMed] [Google Scholar]
- Sugino A., Higgins N. P., Brown P. O., Peebles C. L., Cozzarelli N. R. Energy coupling in DNA gyrase and the mechanism of action of novobiocin. Proc Natl Acad Sci U S A. 1978 Oct;75(10):4838–4842. doi: 10.1073/pnas.75.10.4838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Summers A. O. Untwist and shout: a heavy metal-responsive transcriptional regulator. J Bacteriol. 1992 May;174(10):3097–3101. doi: 10.1128/jb.174.10.3097-3101.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh C. T., Distefano M. D., Moore M. J., Shewchuk L. M., Verdine G. L. Molecular basis of bacterial resistance to organomercurial and inorganic mercuric salts. FASEB J. 1988 Feb;2(2):124–130. doi: 10.1096/fasebj.2.2.3277886. [DOI] [PubMed] [Google Scholar]