Abstract
Expansion of trinucleotide repeats (TNRs) is the causative mutation in several human genetic diseases. Expanded TNR tracts are both unstable (changing in length) and fragile (displaying an increased propensity to break). We have investigated the relationship between fidelity of lagging-strand replication and both stability and fragility of TNRs. We devised a new yeast artificial chromomosme (YAC)-based assay for chromosome breakage to analyze fragility of CAG/CTG tracts in mutants deficient for proteins involved in lagging-strand replication: Fen1/Rad27, an endo/exonuclease involved in Okazaki fragment maturation, the nuclease/helicase Dna2, RNase HI, DNA ligase, polymerase δ, and primase. We found that deletion of RAD27 caused a large increase in breakage of short and long CAG/CTG tracts, and defects in DNA ligase and primase increased breakage of long tracts. We also found a correlation between mutations that increase CAG/CTG tract breakage and those that increase repeat expansion. These results suggest that processes that generate strand breaks, such as faulty Okazaki fragment processing or DNA repair, are an important source of TNR expansions.
Expansion of a CAG/CTG trinucleotide repeat (TNR) causes a number of genetically inherited diseases, including Huntington's disease, myotonic dystrophy, and multiple subtypes of spinocerebellar ataxia (19, 73). By inserting long triplet repeats into the genomes of model organisms, such as bacteria, yeast, and mice, multiple factors have been identified that affect repeat stability. cis-acting factors include the length and purity of a tract, the direction of replication through the tract, its distance from a replication origin, and chromosomal context (16, 17, 48, 51). In addition, several trans-acting factors have been identified whose mutation affects triplet repeat instability, such as the SbcC and Mre11 nucleases in Escherichia coli and yeast, respectively (71, 76); the mismatch repair protein Msh2 in mice (47, 56, 90); and the Rad27 nuclease in yeast (25, 79, 84).
Thus far, all of the triplet repeats whose expansion has been found to cause human disease have an inherent ability to form secondary structures, such as hairpins (CAG, CTG, CGG, and CCG repeats), G quartets (CGG repeats), and triplexes (GAA and CTT) (reviewed in references 61, 65, and 82). These abilities suggest that the primary reason for tract instability is that these secondary structures can interfere with normal cellular processes, such as replication or transcription. Thus, the secondary structures can be viewed as a special category of DNA damage that must be repaired by the cell. CTG repeats form more stable hairpins than CAG repeats in vitro (27, 61). These data are consistent with in vivo observations that show an orientation effect to stability in model organisms: when the CTG strand is on the Okazaki fragment, repeats are more prone to expansions, and CTG repeats on the lagging-strand template are more prone to contractions (26, 42, 57, 60)
In yeast, loss of Rad27, the homolog of human flap endonuclease 1 (Fen1), causes a dramatic increase in expansions of CAG/CTG repeats (25, 79, 84). Fen1/Rad27 has both 5′-3′ exonuclease activity and an endonuclease activity specific for 5′ flap structures (reviewed in references 9 and 43). The in vitro activities and in vivo phenotypes of Fen1/Rad27 indicate that it has an important role in Okazaki fragment processing. Fen1/Rad27 interacts with PCNA (proliferating cell nuclear antigen), the “sliding clamp” that acts as a polymerase processivity factor, and is required for in vitro maturation of Okazaki fragments both in simian virus 40 and yeast replication systems (3, 28, 35). Deletion of RAD27 in yeast causes a replication defect, an increase in mutation frequency, and accumulation of single-stranded DNA at telomeres (64, 69, 83). In addition to triplet repeat changes, other types of mutations accumulate in yeast rad27Δ strains, including duplications flanked by direct repeats and instability of dinucleotide repeats and minisatellite sequences (41, 46, 53, 55, 87). One mechanism that can account for the types of mutations observed in rad27Δ strains is that unprocessed Okazaki 5′ flaps accumulate in the absence of the Fen1/Rad27 protein. When this happens within a TNR tract, fold-back of the flap into a hairpin could further prevent processing and lead to expansions. The phenotype of frequent expansions of CAG/CTG repeats in the absence of Fen1/Rad27 also suggests that defective flap processing is a plausible mechanism for the occasional triplet expansions in the wild-type situation (25, 29, 79). This model is supported by the observation that substrates with secondary structures in the flap are poor substrates for cleavage by Fen1 in vitro (32, 84). In addition, Fen1/Rad27 is essential for long-patch base excision repair (BER) in vitro (44, 45, 81), and rad27Δ strains are very sensitive to methyl methanesulfonate, which creates DNA lesions that are normally repaired by BER (40, 69, 87).
During lagging-strand processing, Fen1/Rad27 works in conjunction with other replication/repair proteins. Fen1 binds to the face of PCNA through a conserved C-terminal PCNA-interacting domain (35). Other replication proteins that bind to PCNA include polymerase δ (Pol δ), the polymerase thought to take over from Pol α after lagging-strand initiation, and DNA ligase. Certain alleles of both PCNA and DNA ligase increase the instability of CAG tracts, although PCNA has the more dramatic effect (36, 78). The Dna2 protein may work in conjunction with Rad27 in maturation of Okazaki fragment 5′ ends. Dna2 is an endonuclease that prefers 5′ single-stranded tails and a 5′-3′ helicase (5, 7, 13). It interacts with Rad27 both genetically and biochemically and is essential for cell viability in yeast (13, 14, 50). Recent characterization of Dna2 activity on model flap substrates led to a two-step model for Okazaki fragment processing (3, 4, 7). In this model, Pol δ displaces a 5′ flap. If the flap becomes longer than ∼27 nucleotides, it is bound by the single-strand binding protein RPA and then cleaved by Dna2, whose endonuclease is stimulated by RPA (4). This cleavage creates a shorter 5- to 7-nucleotide flap that is too short to be bound by RPA but that is a substrate for Fen1/Rad27. In addition, RNase HI may be involved in Okazaki fragment processing, since in vitro it preferentially cleaves 5′ to the last ribonucleotide of an RNA-DNA junction to remove an RNA primer (9, 62). Rnh35, the catalytic subunit of the yeast homolog of mammalian RNase HI, may work cooperatively with Rad27 to remove RNA primers, since Rad27 preferentially cleaves a single-stranded flap of RNA or DNA, whereas yeast RNase H (35) can cleave RNA hybridized to DNA (24, 68). Unlike a rad27Δ strain, rhn35Δ cells have a normal growth rate. However, an rhn35Δ rad27Δ strain has an even slower growth rate and higher mutation rate than a rad27Δ strain, suggesting that the two enzymes may cooperate in Okazaki fragment RNA primer removal (68).
In addition to instability, expanded TNR sequences can induce chromosome fragility. Expanded CCG/CGG tracts at the FRAXA locus and several other loci on human chromosomes show up as gaps or breaks on metaphase chromosomes when cells are grown under conditions that reduce nucleotide pools (63, 85). Both long CCG/CGG and CAG/CTG tracts increase chromosomal breakage at or very near the expanded repeat on yeast chromosomes as well (8, 25). There are several possibilities to explain triplet repeat fragility. First, CCG/CGG and CAG/CTG tracts cause replication fork pausing in E. coli and yeast (66, 75), perhaps because they form secondary structures in vivo, and these stalled forks are likely prone to breakage (58, 59, 74). Second, if damage occurs within a repeat tract, either during replication or unrelated to replication, breakage could occur during repair. For example, a normal intermediate in the repair pathway could get “stuck” because of secondary structure formation by the repeat sequence, leading to an unrepaired break. Fragility of triplet repeats could also play a role in length instability, since several experiments have documented expansions and contractions associated with recombinational repair (25, 37, 70, 71). CAG/CTG repeats also show increased fragility during yeast meiosis that correlates with an increase in meiotic instability (18, 38, 39, 80). Proteins that affect TNR fragility could either increase the initial damage that leads to breakage or be involved in sensing and/or repair of damage. Therefore, identification of such proteins should give insight into what type of repeat-dependent structures are produced in cells and what pathways are used to repair triplet repeat-induced damage.
We developed a novel and sensitive genetic assay that can detect breakage of a CAG/CTG triplet repeat. The assay utilizes a yeast artifical chromosome (YAC) that contains a CAG/CTG repeat tract, several marker genes, and a way to rescue YACs that break at the repeat tract by telomere addition. We used this assay, along with physical analysis of repeat stability, to investigate the effects of mutation of genes coding for proteins involved in lagging-strand replication, including Rad27 (rad27Δ), Dna2 (dna2-1), RNase HI (rnh35Δ), Cdc9 ligase (cdc9-2), Pol δ (pol3-t), and primase (pri2-1), on both the stability and fragility of a CAG/CTG triplet repeat and the relationship between the two phenomena. We show for the first time that loss of RAD27 dramatically increases CAG/CTG fragility. Long CAG/CTG tracts are also more fragile in strains defective in DNA ligase and primase. Strains that show increased fragility also show an increased frequency of TNR expansions. We propose that TNR sequences cause DNA strand breaks by inhibition of correct Okazaki fragment maturation, and repair of breaks within the repeat tract accounts, at least in part, for their propensity to expand.
MATERIALS AND METHODS
Yeast strains and YACs.
All yeast strains were the VPS105 background (MATα ade2 can1 leu2-3,112 Δtrp1 ade3 Δura3 lys2-801 amber) except for cdc9-2 [ATCC 208559; MATα ade2-1 leu2-3 leu2-112 trp1-289 ura3-52 gal2 CUP(r) cdc9-2]. RAD27 was deleted with pMRrad27Δtrp (69). RNH35 was replaced with the TRP1 gene by PCR-based deletion (10). Mutant alleles were introduced via two-step gene replacement with plamids pRS306-dna2-1 (12), YipA16 (pri2-1) (23), and p171 (pol3-t) (88). YACs with various CAG tract lengths were made by modification of YAC VS5 (77) by integration of the CAG tract between the C4A4 and URA3 sequences. This was accomplished by cloning the PvuII fragment from pGEM(CTG)130 containing a CAG tract and 524-bp flanking plasmid DNA (358 bp plus 166 bp) into the blunted NsiI site of pVS20 to create plasmid pCFN-2. pCFN-2 (which contains a URA3 gene) was linearized with AatII and transformed into a wild-type yeast strain containing a Ura3− derivative of YAC VS5, and Ura3+ transformants were selected. The structure of the YAC was confirmed by Southern blot analysis. CAG tract lengths on each YAC were determined by sizing of PCR products on denaturing polyacrylamide gels, on nondenaturing Metaphor gels (FMC Bioproducts; also described below) and by sequencing. YACs were kar crossed into the various mutant backgrounds.
CAG stability and fragility assays.
Cells of each strain were plated for single colonies on YC-Ura-Leu low-adenine plates at the permissive temperature (30°C for the wild-type and rnh35Δ mutant, 23 to 25°C for the other mutants). Pink colonies, indicating one copy of the YAC, were chosen as the starting colonies for the stability and fragility assays. CAG tract length was checked at the start of the experiment by amplification of the CAG tract from colonies by using a small amount of cells as the template (“colony PCR”), primers CTG rev2 (CCCAGGCCTCCAGTTTGC) and T7 (TAATACGACTCACTATAGGG), and Clontech Advantage-GC-Genomic kits in 12.5-μl reactions. Reactions were run on 2% Metaphor gels for 1.5 h at ∼80 V, and the CAG tract size was estimated by comparison with DNA Marker VI (Roche).
For stability assays, three starting colonies with the correct tract lengths were used to inoculate cultures that were grown to approximately three doublings at the semipermissive temperature (30°C). Cells were plated for individual colonies on YC-Ura-Leu plates and incubated at the permissive temperature, and CAG tract length of 48 to 100 individual colonies was analyzed by colony PCR (described above). CAG repeat sizes are accurate to within ∼10 bp or ∼3 repeats for CAG-45 and -70 tracts and within ∼15 bp or ∼5 repeats for CAG-155. The assay was repeated at least three times for each strain and each tract length. Significance was determined by both the Fisher's exact test and a test of equality of binomial proportions for two groups, using Stat Xact 5 software. Ideally, tract length changes should be minimal during growth of these daughter colonies at the permissive temperature, so that any change in tract length reflects an event that happened during growth at the semipermissive temperature. In general, we found this to be true. In the few cases in which only a small proportion of the PCR product ran at a new size (less than ∼50% for contractions and less than ∼20% for expansions, because expanded tracts amplify with less efficiency than smaller tracts), we attributed this to a tract-length change during colony growth (thus, only some cells in the colony would have the new tract length) and did not count this event in the totals.
For fragility assays, 10 starting colonies with correct tract lengths were used to inoculate cultures that were grown to approximately three doublings at the semipermissive temperature (30°C) in YC-Leu (to maintain selection for the YAC but allow loss of the right arm). One hundred microliters of each culture was plated on 5-fluoroorotic acid (FOA)-Leu to select for breakage events, a portion of each culture was combined, and the mixture was plated for single colonies on YC-Leu for a total cell count. Colonies were grown at the permissive temperature and counted, and rates of FOA resistance (FOAR) were calculated by the method of the median (49). Significance was determined by analysis of variance with a post hoc Bonferroni adjustment and by a pooled variance t test, using Systat software.
Analysis of FOAR YAC structure.
FOAR colonies were grown 16 to 24 h in YC-Leu or yeast extract-peptone-dextrose (YEPD) medium, and genomic DNA was isolated by the glass bead method. Genomic DNA was digested with BstEII (New England Biolabs), separated on a 1% agarose gel, probed with a digoxigenin-labeled probe of lambda DNA digested with HindIII, and detected by a chemiluminescent or colorimetric system (Roche). In some cases, a PvuII digest and pYIP5 probe were used to detect only the right terminal fragment of the YAC. Yeast telomere addition to the C4A4 sequence was confirmed by amplification of the junction sequence by using primers CAX29 (CGGCYCGAGCACCCACACCACACCCACAC) and C4A4YIP5 (ATCATTACGACCGAGATTCC), cloning the resulting PCR products, and sequence analysis of clones. Sequencing of one of the lambda healings (wild-type CAG-45, see Fig. 2, lane 10) with primers CAX29 and lambda TG-1690 (TCTGATTAGCCAGGTAACAC) confirmed addition of a yeast telomeric sequence onto a 9-bp telomere-like region of lambda DNA (TGTGGGGTG) present in the last BstEII fragment (see also reference 77).
FIG. 2.
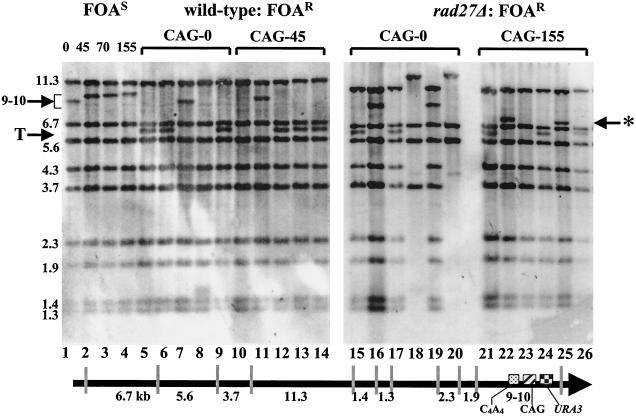
Southern blot analysis of YACs from FOAR colonies. Genomic DNA from FOAS starting colonies (lanes 1 to 4) or FOAR colonies from either wild-type (lanes 5 to 14) or rad27Δ (lanes 15 to 26) strains was purified, digested with BstEII, and separated on a 1% agarose gel. The YAC was visualized by hybridization to a probe to lambda DNA, which makes up the majority of the right arm of the YAC. BstEII restriction fragment sizes (in kilobases) predicted to hybridize to the probe are shown in a diagram at the bottom (not to scale) and to the left of the blot. The last BstEII fragment contains the backup telomere (C4A4), CAG tract, and URA3 gene, represented by boxes, and is predicted to be between 9 and 10 kb, depending on the size of the CAG tract (indicated by an arrow to the left of the gel). Breakage at the tip of the right arm—for example, at the CAG tract—and healing of the YAC by telomere addition at the backup telomere will reduce the size of the teminal band to approximately 6.6 kb (T). Healing at the CAG tract is expected to give a 7- to 7.5-kb terminal band (*). Healing behind the backup telomere sequence in the lambda sequence will result in loss of one or more of the bands, sometimes with appearance of new bands.
RESULTS
A novel YAC-based assay to monitor stability and fragility of TNRs.
CAG/CTG tracts have been shown to cause an increase in chromosome breakage in yeast in a tract-specific manner by both genetic and physical assays (25). The genetic assay used to monitor CAG/CTG-tract breakage as described by Freudenreich et al. (25) was based on placing a CAG/CTG repeat tract on a natural yeast chromosome next to a URA3 marker gene and between two direct repeats. A breakage event at the CAG/CTG tract could be healed by degradation of both the tract and the URA3 gene to expose the direct repeats, followed by single-strand annealing. These recombination events can be identified because cells that express Ura3p die in the presence of the drug FOA, whereas cells that lose the URA3 gene are FOAR. Therefore, only recombination-proficient cells are able to recover the broken chromosome.
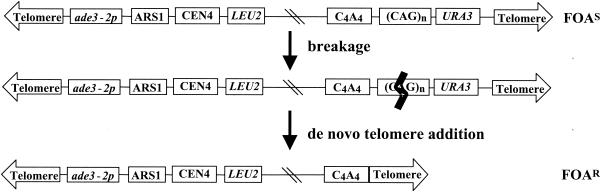
Because many proteins involved in DNA replication are also necessary for efficient recombination, we devised an alternative assay based on a YAC (YAC-CF1; Fig. 1) that does not require recombination (77). In this assay, CAG/CTG tracts of various lengths (referred to as CAG) were placed on the right arm of a YAC, proximal to a URA3 gene. The tract is oriented such that the CAG strand will be on the lagging-strand template and the CTG strand will be on the Okazaki fragment. This orientation is less prone to contractions (26, 57). The URA3 gene on YAC-CF1 is far enough away from the telomere that it is not subject to telomeric silencing, thus cells containing the original YAC are Ura+ and FOA sensitive (FOAS) (Fig. 1) (77). Just proximal to the CAG tract there is a “backup telomere”, a 108-bp stretch of C4A4/T4G4 telomeric sequence from Oxytricha, a sequence that is used efficiently for telomere addition by yeast telomerase (67). If breakage occurs at the CAG tract, the end of the broken YAC is degraded, exposing the C4A4 sequence and allowing it to act as a seed for addition of a yeast telomere. Telomere addition allows the efficient recovery of YACs that break at the CAG tract. Breakage followed by telomere addition leads to loss of the right arm of the YAC, including the URA3 gene, and generates a Ura3− and FOAR cell (Fig. 1). Thus, measurement of the rate of generation of FOAR cells can be used as a monitor of the rate of breakage. Since breakage does not occur in a region with homology to yeast chromosomes, the primary mode of healing is by telomere addition rather than recombination (77). This is supported by the fact that a deletion of the RAD52 gene, required for homologous recombination, does not affect the rate of generation of FOAR cells (R. Sundararajan and C. H. Freudenreich, unpublished data). The left arm of the YAC has LEU2, which is used to maintain selection for the YAC, a yeast origin of replication (ARS1), a centromere (CEN4), and a promoter-defective allele of ADE3 (ade3-2p) that allows determination of YAC copy number by color of the colony (0 YACs = white, 1 YAC = pink, 2 YACs = red).
FIG. 1.
The YAC breakage assay. Cells containing YAC CF1 (top; not to scale), which contains a CAG tract and URA3 gene, are FOAS. If breakage occurs distal to the backup telomere (C4A4 tract)—for example, in the CAG tract (shown by a heavy line)—the YAC can be rescued by addition of a yeast telomere onto the C4A4 seed sequence. The resulting YAC has lost the URA3 gene and is thus FOAR. The YAC is ∼62 kb long and contains 41 kb of lambda DNA between the LEU2 gene and C4A4 sequence.
Along with the genetic assay for fragility, stability of the CAG repeat on the YAC can be monitored by a physical PCR assay, using primers on either side of the repeat to amplify the tract and monitor changes in size (described below for details). The YAC is especially well suited to study how various mutations affect TNRs, since it is a dispensable chromosome and can be moved easily to any strain of interest by cytoduction (22). Thus, both fragility and stability of various repeats cloned on the YAC can be analyzed by simply moving the YAC to different mutant strain backgrounds.
Validation of the YAC assay as a monitor of TNR fragility.
Southern analysis of the structure of a YAC without a CAG tract in FOAR cells generated in a wild-type strain showed that the majority of YACs healed by telomere addition at the C4A4 tract (77). FOAR events were also generated by point mutations in URA3 at a rate of 2 × 10−7 and a small proportion of YACs healed behind the C4A4 tract in the lambda sequence that composes the majority of the YAC (77). The latter two events can be distinguished from de novo telomere addition at the C4A4 tract by determining the structure of the YAC in FOAR cells.
To determine what proportion of FOAR cells was generated by breakage and telomere addition on YAC-CF1 with CAG tracts of different lengths, we carried out restriction digestion and Southern analysis to determine the structure of the YAC in independent FOAR colonies (Fig. 2, left panel, and Table 1). After restriction digestion, the lambda probe hybridizes to a series of fragments derived from the right arm of the YAC. In the starting YAC, the terminal fragment containing the C4A4 sequence, the CAG repeat, and the URA3 gene is ∼9 to 10 kb depending on the CAG repeat size (Fig. 2, arrow, lanes 1 to 4). FOAR due to breakage at the CAG tract would most likely yield a YAC that healed at the backup telomere, since the C4A4 tract, an efficient seed for telomere addition, is located immediately proximal to the CAG tract. If a new telomere was added to the C4A4 tract, the terminal fragment will decrease in size to ∼6.6 kb (Fig. 2, T arrow). Southern analysis showed that the majority of YACs from FOAR cells had healed at the C4A4 sequence, consistent with a breakage event distal to the backup telomere, and the percentage increased with increasing tract length (56% for CAG-45; 75% for CAG-70; 100% for CAG-155; Table 1; see Fig. 2, lanes 5, 6, 9, 12, 13, and 14 for examples). Sequence analysis of two healings (of a CAG-45 and CAG-70 YAC) confirmed that yeast telomeric DNA was added to the C4A4 sequence. In the cases in which the YAC remained full length, FOAR is attributed to a point mutation in the URA3 gene (Fig. 2, lanes 7 and 11; average of 19% for the wild-type strain) (Table 1). In a few cases, the YAC healed internal to C4A4, probably indicative of a random breakage event, since these types of events were more frequent in the CAG-0 and -45 YACs than in YACs with longer CAG tracts (Table 1; i.e., see Fig. 2, lane 8, in which the 9-kb band is gone and a new ∼1.6-kb band appears; this site of healing corresponds to an 11-bp telomere-like sequence in the lambda DNA) (77). Because we observed that the majority of FOAR colonies had a structure consistent with breakage at or near the CAG tract, with the proportion increasing as CAG tract length increased, the YAC assay can be used to measure TNR fragility and to study factors that influence TNR tract breakage.
TABLE 1.
Healing of YACs purified from FOAR colonies as determined by Southern blot analysis
| Strain and tracta | No. of YACs healed atb:
|
% Tel+CAGc | |||
|---|---|---|---|---|---|
| Telomere | Point mutation | Lambda | CAG | ||
| Wild type | |||||
| CTG-0 | 4 | 3 | 2 | 0 | 45 |
| CTG-45 | 5 | 1 | 3 | 0 | 56 |
| CTG-70 | 3 | 1 | 0 | 0 | 75 |
| CTG-155 | 4 | 0 | 0 | 0 | 100 |
| Total (26) | 16 | 5 | 5 | 0 | 62 |
| rad27 | |||||
| CTG-0 | 6 | 2 | 3 | 0 | 55 |
| CTG-45 | 2 | 1 | 4 | 1 | 38 |
| CTG-70 | 4 | 1 | 2 | 1 | 63 |
| CTG-155 | 5 | 1 | 3 | 5 | 71 |
| Total (41) | 17 | 5 | 12 | 7 | 59 |
| dna2-1 | |||||
| CTG-0 | 2 | 1 | 2 | 0 | 40 |
| CTG-45 | 2 | 1 | 2 | 0 | 40 |
| CTG-70 | 3 | 0 | 1 | 0 | 75 |
| CTG-155 | 5 | 0 | 0 | 0 | 100 |
| Total (19) | 12 | 2 | 5 | 0 | 64 |
| rnh35 | |||||
| CTG-0 | 2 | 0 | 2 | 0 | 50 |
| CTG-45 | 2 | 1 | 2 | 0 | 40 |
| CTG-70 | 3 | 1 | 1 | 0 | 60 |
| CTG-155 | 3 | 1 | 0 | 0 | 75 |
| Total (18) | 10 | 3 | 5 | 0 | 56 |
| cdc9-2 | |||||
| CTG-0 | 3 | 1 | 1 | 0 | 60 |
| CTG-45 | 5 | 0 | 0 | 0 | 100 |
| CTG-70 | 5 | 0 | 0 | 0 | 100 |
| CTG-155 | 4 | 1 | 0 | 0 | 80 |
| Total (20) | 18 | 1 | 1 | 0 | 85 |
| pol3-t | |||||
| CTG-0 | 3 | 2 | 0 | 0 | 60 |
| CTG-45 | 3 | 1 | 1 | 0 | 60 |
| CTG-70 | 4 | 0 | 1 | 0 | 80 |
| CTG-155 | 3 | 1 | 0 | 0 | 60 |
| Total (19) | 13 | 4 | 2 | 0 | 68 |
| pri2-1 | |||||
| CTG-0 | 3 | 0 | 1 | 0 | 75 |
| CTG-45 | 3 | 0 | 2 | 0 | 60 |
| CTG-70 | 4 | 0 | 1 | 0 | 80 |
| CTG-155 | 3 | 0 | 2 | 4 | 78 |
| Total (23) | 13 | 0 | 6 | 4 | 74 |
The total number of colonies analyzed for each strain is listed in parentheses.
Telomere, healed at backup telomere (C4A4 sequence); Point mutation, starting-size YAC, putative point mutation in the URA3 gene; Lambda, healed proximal to C4A4 in lambda sequence as assessed by loss of bands; CAG, probable healing in CAG tract.
% Tel+CAG, percent healed at either backup telomere or CAG repeat.
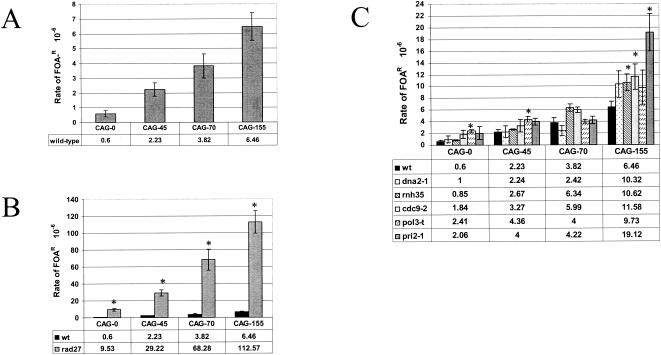
In human cells, CAG tracts of ≤37 repeats are stable and not subject to frequent expansions or contractions, while tracts ≥37 repeats are unstable, with the extent of instability increasing with increasing tract length. To test whether CAG tracts showed length-dependent breakage on YAC-CF1 as they had on yeast chromosome II (25), we determined the rate at which FOAR colonies were generated in wild-type cells carrying YACs with 0, 45, 70, or 155 CAG repeats (CAG-0, -45, -70, or -155, respectively). The results show that the rate of FOAR increases ∼11-fold as the tract length increases from 0 to 155 repeats (Fig. 3A). This increase is similar to the increased breakage rate on chromosome II for CAG tracts of similar length (25), indicating that expanded CAG tracts integrated in YAC-CF1 also act as preferred sites of breakage in a length-dependent manner.
FIG. 3.
Breakage assay of YAC-CF1, containing CAG-0, -45, -70, or -155 tracts in different strain backgrounds. For each strain, 10 colonies of each tract length were grown separately for approximately three doublings at 30°C (which is semipermissive for the temperature-sensitive strains used) in Ura+ Leu− medium to allow for breakage and healing at the backup telomere and then plated on FOA-Leu plates to select for breakage events and YC-Leu medium for a total cell count, and grown at the permissive temperature for each strain (23 or 25°C for temperature-sensitive mutants, 30°C for rnh35Δ and wild-type strains). Rates of FOAR (10−6) were calculated by the method of the median (49). Rates that are significantly different from those of the wild type by a pooled variance t test are shown by asterisks (P < 0.05); standard errors are shown. (A) Wild-type strain. (B) Comparison of wild-type and rad27Δ strains. (C) Comparison of the wild-type strain and strains with mutations of other proteins involved in lagging-strand synthesis. Note differences in the scales for each graph.
Mutation of a subset of proteins involved in lagging-strand replication, including a deletion in RAD27, increases CAG/CTG tract breakage.
Since a defect in processing the 5′ end of Okazaki fragments in a rad27Δ strain leads to a large increase in CAG/CTG repeat expansions, we were interested in testing whether there was increased fragility of the tract in a rad27Δ strain background. rad27Δ strains show a conditional lethality at 37°C and grow more slowly than the wild type at 30°C (69, 83). rad27Δ strains containing YACs with 0, −45, −70, or −155 repeats were plated on YC-Ura-Leu plates to maintain selection for both arms of the YAC and incubated at the permissive temperature (23°C) to form individual colonies. Colony PCR was done to find 10 colonies of each tract length that had the correct starting CAG repeat size. For each tract length, the 10 colonies were used to inoculate individual YC-Leu cultures; this medium selects for the left arm of the YAC, but allows loss of the right arm containing the URA3 gene. Cultures were grown at 30°C (a semipermissive temperature) for approximately three doublings to allow for breakage and healing of the YAC. A portion of each culture was plated on FOA-Leu plates to select FOAR colonies, and another portion was plated for a total cell count. The rate of breakage was determined by the method of the median (49).
The YAC breakage assay revealed that for YACs containing a CAG repeat, the rad27Δ strain gave a dramatic increase in the rates of generating FOAR cells (13-fold increase compared to the wild type for CAG-45, 18-fold for CAG-70, and 17.4-fold for CAG-155; Fig. 3B). The background rate of FOAR in the CAG-0 strain was also elevated compared to that of the wild type, indicating that overall chromosome breakage is increased in the absence of the Rad27 protein. However, there was also a marked tract-specific effect with higher rates of FOAR as the repeat tract increased in length, and the rates of FOAR at the higher tract lengths were much greater than expected for a simple additive effect of the background increase in FOAR due to the mutation plus the increase in FOAR due to tract length seen in the wild-type background. For example, if the rad27Δ CAG-0 value and the wild-type CAG-70 value are both subtracted from the rad27Δ CAG-70 rate, the rate of FOAR is still 54.93 × 10−6, compared to 3.82 × 10−6 for the wild type. These data suggest that the propensity for long CAG tracts to break is specifically exacerbated in the rad27Δ background.
Because Rad27 is involved in Okazaki fragment maturation, we analyzed other proteins involved in lagging-strand replication and processing to monitor their effects on CAG fragility. Mutations in three proteins that are thought to work together with Rad27 in processing the 5′ ends of Okazaki fragments were tested: the Dna2 helicase/nuclease, RNase H, and DNA ligase. The dna2-1 mutant is partially defective in both the helicase and nuclease activities (14) and grows more poorly at increasing temperatures (12, 13). We maintained the strain at 23°C, a permissive temperature with almost wild-type DNA synthesis, and allowed breakage at the semipermissive temperature of 30°C. RNase HI activity, thought to be involved in RNA primer removal, is reduced by 75% by a deletion in the yeast RNH35 gene, but the growth rate of the deletion mutant is essentially wild type at 30°C (24, 68). DNA ligase is required for the last step of Okazaki fragment processing and ligation of the processed fragment to the upstream fragment. A strain carrying the cdc9-2 ligase mutation arrests as large budded cells at 37°C and shows reduced viability at the semipermissive temperature of 30°C, indicating a defect in DNA replication (31). In vitro assays using cdc9-2 extracts showed a complete defect in ligation during base and nucleotide excision repair (NER) pathways at 30°C (91).
None of the three processing mutants, dna2-1, rnh35Δ, or cdc9-2 significantly increased FOAR for the CAG-0, -45 or -70 YACs, although the rnh35Δ and cdc9-2 strains showed a slight increase for CAG-70 (Fig. 3C). However, all three mutants did show an increase in FOAR for strains with the CAG-155 YAC, with the rnh35Δ and cdc9-2 rates statistically significant compared to the wild-type rate. All three CAG-155 rates were still elevated if the respective no-tract control and wild-type CAG-155 rates were subtracted out. Thus, elimination or decrease of either RNase HI or ligase activity, and perhaps also Dna2-1 activity, appears to have an effect on the breakage of long CAG tracts.
Mutations in two proteins involved in generation of Okazaki fragments, Pol δ and DNA primase, were also tested for effects on CAG fragility. The pol3-t allele is a D641N substitution in the vicinity of the conserved polymerase motif VI of Pol δ (88). This allele increases the mutation rate by a number of different assays, but has the most dramatic effect on deletions between distant short repeats (88, 89). The pol3-t allele significantly increased FOAR of the CAG-0 and CAG-45 YACs, but not CAG-70 or CAG-155 (Fig. 3C), suggesting that this allele may increase overall chromosome breakage slightly, but does not have a tract-specific effect.
The pri2-1 allele is an E152Q mutation in the catalytic subunit of DNA primase that causes cells to cease DNA synthesis and arrest at 37°C (23). Interestingly, the pri2-1 allele had no effect on breakage of the shorter CAG-45 and -70 tract lengths, but showed a threefold increase in FOAR compared to the wild type for the CAG-155 tract (Fig. 3C, P = 0.003 by t test). The increase was still significant if the respective no-tract control and wild-type CAG-155 rates were subtracted out. This allele also increased expansions of the CAG-155 tract by ∼11-fold (but not of CAG-45 or -70, described below).
Southern analysis of DNA from FOAR colonies shows that rates of FOAR also reflect CAG/CTG tract breakage in mutant backgrounds.
To verify that the rate of FOAR reflected breakage at the CAG tract in the mutant strains, DNA was purified from at least four FOAR colonies for each tract length (CAG-0, -45, -70, and -155) for each mutant background and the structure of the YAC was examined by Southern blot analysis (Fig. 2 and Table 1). Except for the rad27Δ and pri2-1 strains (described below), each of the mutants showed a similar frequency of healing at the backup telomere (average of 67% for all YACs analyzed) with the frequency generally increasing as CAG tract length increased (Table 1). If the rates of FOAR shown in Fig. 3B are corrected so that only telomere healings at the C4A4 tract are considered, the rad27Δ strain still shows dramatic and highly significant increases over the wild type at all CAG tract lengths (range, 9- to 13-fold). We infer from these data that the rate of FOAR is a good approximation of the rate of CAG tract breakage.
In two cases, less-than-average healing at the backup telomere was observed. For the rad27Δ strain, only 41.5% of YACs healed at the backup telomere (17 out of 41 rad27Δ FOAR colonies analyzed by Southern blotting) (Table 1). This lower frequency was almost entirely due to a new site of telomere addition, because the size of the most-distal YAC fragment predicted that telomere addition had occurred at the CAG repeat tract rather than at the C4A4 tract (see Fig. 2, lanes 22 and 25; and Table 1, CAG). This type of healing did not occur in the rad27Δ CAG-0 strain (0 of 11 FOAR colonies), infrequently in CAG-45 and -70 (1 of 8 FOAR colonies each), but was observed frequently for the CAG-155 repeat (5 of 14 FOAR colonies). The only other strain that showed frequent telomere addition within the CAG tract was the pri2-1 CAG-155 strain, in which four of nine FOAR colonies analyzed had healed in the CAG tract (Table 1). If these potential CAG healing events are added to the backup telomere healings on the premise that they most likely arise from breakage at the CAG tract, then the frequencies of FOAR colonies that can be attributed to CAG tract breakage are 71% for rad27Δ CAG-155 and 78% for pri2-1 CAG-155, which is close to the average of 83% seen for the CAG-155 tract in the other strain backgrounds. Although a deletion in RAD27 has been shown to increase gross chromosomal rearrangements, including translocations, we did not see any direct evidence for these structures (15). However, the rad27Δ strain did have a fairly high frequency of loss of bands in the lambda sequence (Table 1; Fig. 2, lanes 18 and 20), and some of these might represent translocation events.
Because telomere structure is affected in a rad27Δ strain and we observed an altered spectrum of telomere healings in this strain, we were concerned that rescue of broken YACs by telomere addition might be less efficient than in the wild-type strain, masking a real rate of CAG breakage even higher than the observed 13- to 18-fold increase. To address this possibility, we determined the rate of CAG tract breakage by using our previously described direct repeat recombination assay, which depends on single-strand annealing but not telomere addition (25). With this assay, there was a 15- to 18-fold increase in FOAR for a CAG-80 strain, and there was a 10- to 11-fold increase for a CAG-100 strain compared to the wild-type controls (data not shown). Since these increases were similar to those seen in the YAC assay, it is unlikely that inefficient telomere healing had a major effect on the rates observed. In addition, telomere structure was checked by Southern blotting for the two mutants that did not show significant increases in CAG-155 breakage rates (dna2-1 and pol3-t), and in both cases, telomeres appeared intact at the 30°C semipermissive temperature (although slightly longer than wild type for dna2-1) (data not shown), suggesting that telomere structure and, by inference, telomere healing were not grossly altered in these strains.
Mutations that increase fragility of CAG/CTG tracts also increase tract expansions.
In order to determine if fragility correlates with stability of the CAG tracts, we also tested the frequency of contractions and expansions that occurred under the same growth conditions used for the YAC breakage assay. Individual yeast colonies of either a wild-type or mutant strain background containing the indicated YAC were grown at the permissive temperature (30°C for wild type and rnh35Δ, 23 to 25°C for temperature-sensitive mutant strains rad27Δ dna2-1 cdc9-2 pol3-t pri2-1 strains), and the CAG tract length was analyzed by colony PCR to find colonies that contained full-length tracts. These parent colonies were then used to inoculate a culture that was grown for approximately three doublings at the semipermissive temperature (30°C). During this period of growth, the CAG tract should be replicated under conditions of reduced or absent protein product, so that any effects on tract stability would be manifest. A dilution was then plated for single cells that were allowed to grow into colonies at the permissive temperature, and the CAG tract size of individual colonies was determined by colony PCR (Table 2). Because PCR preferentially amplifies short tracts and we were able to amplify a maximum of 180 to 200 repeats by PCR, the CAG-155 expansion sizes and frequencies were probably underestimated.
TABLE 2.
Frequency of CAG/CTG expansions and contractions in wild-type and mutant strain backgrounds
| Strain backgrounda | % Contraction (fold)b
|
% Expansion (fold)b
|
||||
|---|---|---|---|---|---|---|
| CAG-45 | CAG-70 | CAG-155 | CAG-45 | CAG-70 | CAG-155 | |
| Wild type | 0.8 (1) | 3.7 (1) | 16.8 (1) | 0.4 (1) | 0.8 (1) | 0.5 (1) |
| rad27Δ | 4.0 (4.9)* | 13.3 (3.6)** | 40.3 (2.4)** | 11.0 (26.8)** | 27.7 (33.8)** | 10.4 (20)** |
| dna2-1 | 2.8 (3.4) | 6.9 (1.9) | 25.0 (1.5) | 1.4 (3.4) | 2.1 (2.5) | 0 (0) |
| rnh35Δ | 0.7 (0.8) | 4.0 (1.1) | 23.6 (1.4) | 0.7 (1.6) | 2.0 (2.4) | 1.39 (2.7) |
| cdc9-2 | 0 (0) | 4.2 (1.1) | 25.0 (1.5) | 0.7 (1.7) | 2.1 (2.5) | 4.9 (9.3)* |
| pol3-t | 2.1 (2.5) | 11.8 (3.2)** | 62.6 (3.7)** | 2.1 (5.0) | 0 (0) | 2.0 (3.9) |
| pri2-1 | 4.2 (5.0)† | 10.4 (2.8)* | 29.9 (1.8)** | 0 (0) | 1.4 (1.7) | 5.6 (10.7)** |
A minimum of 144 to 150 samples were evaluated for each tract length of each strain. For wild-type and rad27Δ strains, the following numbers of samples were tested: wild-type CAG-45, 241; CAG-70, 245; CAG-155, 191; rad27Δ CAG-45, 200; CAG-70, 195; and CAG-155, 144.
Frequencies are expressed as percentages; fold increases over wild-type are indicated in parentheses. **, P <0.01; *, P <0.05; †, P <0.057 (Fisher's exact test and a test of equality of binomial proportions).
In the wild-type strain background, the frequency of CAG tract expansions was very low (0.4 to 0.8%, Table 2). The frequency of contractions was also low for the short CAG-45 tract (0.8%), but as in human transmissions, tract instability increased significantly for longer repeat tracts (3.7% contractions for CAG-70, 16.8% for CAG-155) (Table 2) As has been noted before (26, 57, 60), contractions were more frequent than expansions in wild-type yeast.
Consistent with earlier studies, repeat instability was dramatically higher in a rad27Δ strain, with a strong bias toward expansions (2- to 5-fold increase in contractions and 20- to 34-fold increase in expansions compared to that in the wild type) (Table 2). The dna2-1 defect slightly increased contractions for all tract lengths with a 3.4-fold increase for the shorter CAG-45 tract and also increased expansions for the CAG-45 (3.4-fold) and CAG-70 tracts (2.5-fold) (Table 2). The increases were modest and not statistically significant by a Fisher's exact test, suggesting that mutation of the Dna2 protein alone did not have a dramatic effect on CAG stability, but might have a modest effect for shorter repeat tracts. The rnh35Δ did not affect CAG contractions, but led to a slight increase in expansions for the longer CAG-70 and -155 tracts (2.4- to 2.7-fold [not statistically significant]) (Table 2). The cdc9-2 mutation had a mild effect on CAG expansion of shorter tracts and showed a significant ninefold increase in expansions of the longest CAG-155 tract, the same tract length that showed increased fragility.
Consistent with its previously characterized phenotype on direct repeats and dinucleotide repeats (46, 89), the pol3-t mutation significantly increased contractions of both CAG-70 and -155 tracts. The mutation also increased expansions for the CAG-45 and -155 tracts, although the values were not statistically significant, suggesting a minor effect on repeat expansions, but, similar to pol3-t-induced fragility, the effect may not be dependent on repeat tract length. Like the pol3-t mutation, the pri2-1 allele increased contractions of CAG repeats. In addition, there was a large 11-fold increase in frequency of expansions for the longest CAG-155 tract in a pri2-1 strain, again the same length that showed increased fragility.
We also analyzed the mean and the range of the tract length changes seen in wild-type cells and all of the mutant backgrounds (Table 3). Most of the contractions detected in wild-type cells were fairly large, so that the average contracted tract was only about half the size of the original tract or less. None of the mutations tested had a noteworthy effect on the size of tract contractions. The mean size of expansions in the wild-type background was slightly less than the contractions, but still fairly large (25 repeats for CAG-45, 30 repeats for CAG-70, 15 repeats for CAG-155). Again, most mutations did not affect the size of expansions, with one exception. For the rad27Δ strain, we saw several large expansions of the CAG-70 repeat that doubled or almost doubled the size of the tract (60 to 70 repeats added), whereas the largest documented expansions for all other strains were additions of 30 to 45 repeats. The largest was an expansion of ∼100 repeats (165 repeats total). Such large expansions were not observed for the CAG-45 and -155 tracts in the rad27Δ strain; however, for the CAG-155 tract, we had a high proportion of failed reactions (26% of reactions versus 0% for wild-type CAG-155), indicating that technical limitations of the PCR likely led to an underestimation of expansion sizes. These results suggest a possible mechanistic difference between expansion of shorter CAG-45 and longer CAG-70 tracts.
TABLE 3.
Size of CAG/CTG tract-length changes observed in stability assays
| Strain | No. of repeatsa
|
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CAG-45
|
CAG-70
|
CAG-155
|
||||||||||
| Contractions
|
Expansions
|
Contractions
|
Expansions
|
Contractions
|
Expansions
|
|||||||
| Range | Mean | Range | Mean | Range | Mean | Range | Mean | Range | Mean | Range | Mean | |
| Wild type | 20-40 | 30 | 25 | 25 | 10-60 | 35 | 25-35 | 30 | 25-130 | 70 | 15 | 15 |
| rad27Δ | 10-40 | 20 | 5-30 | 25 | 5-70 | 25 | 10-100 | 35 | 25-140 | 70 | 5-10 | 10 |
| dna2-1 | 10-15 | 10 | 10-15 | 10 | 10-60 | 30 | 15-40 | 30 | 25-130 | 70 | — | — |
| rnh35Δ | 15 | 15 | 30 | 30 | 20-50 | 30 | 10-45 | 30 | 30-130 | 80 | 30 | 30 |
| cdc9-2 | — | — | 25 | 25 | 5-50 | 15 | 15-30 | 25 | 20-140 | 75 | 5-10 | 10 |
| pol3-t | 10-20 | 15 | 15-25 | 20 | 5-60 | 30 | — | — | 25-140 | 75 | 5-10 | 10 |
| pri2-1 | 10-30 | 15 | — | — | 5-45 | 20 | 20-40 | 30 | 30-150 | 70 | 10-25 | 15 |
Shown are numbers of CAG/CTG repeats added to (for expansions) or subtracted from (for contractions) the original tract length and rounded to the nearest 5 to reflect the accuracy of the sizing data (i.e., 25 in the CAG-45 expansion column means that ∼25 repeats were added for a total of ∼70 repeats). —, no expansions or contractions observed.
DISCUSSION
It is intriguing that TNR tracts are unstable in two different ways, that is they change in size and also cause chromosome fragility, and both types of instability are exacerbated as the tract increases in length. It has been tempting to speculate that the two types of instability are interrelated, but until now, there has been no direct connection between increased CAG breakage and instability outside of meiosis. We have developed a quick YAC-based genetic assay that does not rely on recombination to monitor the effects of genes and conditions on TNR-induced chromosome fragility. Using this assay, we show that mutation of Fen1/Rad27, DNA ligase, and primase causes increased CAG fragility and that there is a correlation between the level of fragility and the likelihood of repeat expansions. On the other hand, there was no obvious relationship between increased fragility and TNR contractions.
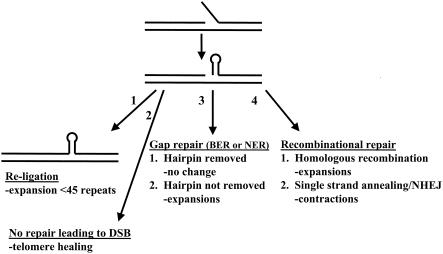
One striking aspect of the fragility data was the large increase in breakage observed in a rad27Δ strain, suggesting that a defect in flap processing leads to chromosomal breakage more often than other types of replication defects. The increase in breakage with increasing tract length indicates that TNRs either exacerbate the initial lesion (i.e., unprocessed flaps), inhibit effective repair of unprocessed flaps, or both. For example, a proportion of CTG-containing flaps could be refractory to ligation, leading eventually to a double-strand break (DSB) that would be a substrate for telomere addition (Fig. 4, pathway 2). Alternatively, repair (Fig. 4, pathways 3 and 4) could be aborted because of unresolved secondary structures, leading to telomere addition instead. Aborted repair could be more common in mutant strains: Rad27 plays a role in long-patch BER and end joining, and DSB repair that involves coordinated leading- and lagging-strand synthesis requires most proteins involved in lagging-strand replication, including Rad27, ligase, and primase (34, 44, 45, 92, 93).
FIG. 4.
Model for TNR expansions and breakage due to hairpin formation at flap structures. Displaced 5′ flaps, formed either on the lagging strand during replication or during nick or gap repair, that contain CTG repeats are predicted to fold into a hairpin structure at some frequency. The likelihood of hairpin formation is increased in strains defective for flap processing, such as strains lacking Fen1/Rad27. Four possible pathways for resolution of the hairpin/nick structure and their consequences for repeat instability are shown. NHEJ, nonhomologous end joining.
A deletion in RAD27 also had a dramatic effect on TNR stability, especially increasing the number of expansions, consistent with earlier results obtained with different assays (25, 79, 84). There are two general mechanisms that could account for the increase in expansions seen in the absence of Rad27 or mutation of ligase or primase (Fig. 4). First, a proportion of unprocessed flaps could eventually be ligated to the preceding Okazaki fragment (Fig. 4, pathway 1). In vitro studies have shown that DNA ligase I can compete with Fen1 during CTG flap processing to create expanded products, and production of expanded products is increased when Fen1 is mutated (33, 52). Additional displacement synthesis by Pol δ could possibly increase the size of the flap even further, perhaps even to the size of the majority of the Okazaki fragment (100 to 150 nucleotides = 33 to 50 repeats) (2, 11). The extra sequence contributed by the unprocessed flap would lead to an expansion, but the maximal size of the repeat tract added would be limited by the size of an Okazaki fragment, or ∼45 repeats, which is the maximum seen in all strains except rad27Δ.
Alternatively, a repeat-containing flap that is unable to be correctly processed could initiate repair, and expansions could occur during repair (Fig. 4, pathways 3 and 4). Loss of the end of the YAC and healing at the backup telomere are fairly low-frequency events and probably only account for a small fraction of breakage events, probably around 1% (77; data not shown). Therefore, most breakage events probably heal either intrachromosomally (pathways 3 and 4.2) or by a homologous recombination pathway using the sister chromatid as a template (pathway 4.1). A gap repair model (pathway 3) has been proposed to explain the CAG expansions that occur at the Huntington's locus in transgenic mouse haploid germ cells (47), and abasic sites have been shown to induce TNR expansions in vitro (54). Recently, polymerase β, which functions in short-patch BER, has been shown to expand CAG/CTG repeats at strand breaks in vitro (30). For example, fill-in synthesis during gap repair combined with the absence of flap cleavage, followed by eventual ligation, could produce TNR expansions.
Different from other mutations, some very large expansions were observed in the rad27Δ background that approximately doubled the size of the CTG/CAG-70 repeat tract. Spiro et al. observed a similar frequency of large expansions in a rad27Δ strain when a CTG-25 tract was on the Okazaki fragment (as in this study) and an even larger fraction in the opposite orientation (CAG-25) (84). Multiple rounds of unprocessed flap religation or replication slippage during gap repair could possibly lead to these large expansions. Alternatively, recombinational repair (Fig. 4, pathway 4) using the sister chromatid as a template could account for generation of the occasional large expansions observed in the rad27Δ strain. One version of recombinational repair that can account for the nonreciprocal nature of TNR expansions is synthesis-dependent strand annealing (see reference 72 for review). The idea that DSB repair can generate expansions is supported by the high rate of CAG tract breakage in the backgrounds that showed expansions, indicating that there are more DSBs arising within or near the repeat compared to that of the wild type. There is good evidence that the lesions generated in a rad27Δ strain are subject to recombinational repair, since a RAD27 deletion is lethal in combination with almost all proteins needed for homologous recombination (20, 86). Recombinational repair can also proceed by single-strand annealing, a pathway that should readily occur in a repeated sequence, or nonhomologous end joining, both of which would lead to contractions. In addition to the large increase in TNR expansions, the rad27Δ strain showed a higher frequency of contractions compared to that of the wild type, consistent with the idea that recombinational repair pathways are increased in the rad27Δ background. The model that DNA strand breaks in a CAG/CTG tract can be an initiator of repeat instability is also supported by earlier experiments that indicated that DSB repair in the vicinity of a repeat tract can initiate repeat instability that is biased toward expansions. First, a recombination event near a CAG tract on a yeast chromosome led to the recovery of large expansions (25). Second, repair of an induced DSB by using a CAG/CTG repeat template increases both expansions and contractions over the normal mitotic rate in yeast (70, 71). In addition, RecA-dependent recombination between CAG/CTG repeats in E. coli also produces a bias toward expansions, whereas there is usually a bias toward contractions during replication of CAG-containing plasmids in bacteria (37). Finally, CAG repeats also act as sites of chromosome breakage in yeast meiosis, and meiotic instability of CAG repeats has recently been shown to depend on creation of the DSB by the SPO11 endonuclease (38, 39).
In contrast to the Rad27 deletion, mutation of three other proteins thought to cooperate with Rad27 in flap processing, Dna2, RNase HI, and ligase had much more subtle effects on CAG fragility and stability. Although the Dna2 and ligase alleles must retain some function at the semipermissive temperature used, since a deletion of each of these proteins is lethal, both show reduced or no activity in vitro and significantly reduced viability at 30°C; thus, any strong effects on TNRs should have been revealed. The cdc9-2 ligase-defective strain showed a significant increase in both fragility and expansions of the longest CAG-155 tract. This result supports the idea that the persistence or repair of unligated nicks on the lagging strand is one pathway to expansions. Expansions of the CAG-70 tract were slightly less frequent than that observed previously for a CAG-78 tract in either a cdc9-1 or cdc9-2 mutant background (36). The basis for this difference is not known, but could be due to a difference in strain background or chromosomal location.
The lack of any significant effects of the dna2-1 mutation on fragility and stability of CAG tracts indicates that Dna2 is probably not required in vivo for maturation of CTG-containing flaps in the presence of wild-type Rad27 protein and suggests that most flaps are shorter than 28 nucleotides (a size that no longer supports efficient loading and cleavage by Fen1/Rad27 due to competition by RPA) (4, 7). This conclusion is in agreement with the data of Ayyagari et al. that show an average nick translation patch of 8 to 12 nucleotides in vitro (3). However, a slight increase in fragility of the longest CAG-155 tract was noted in the presence of the dna2-1 mutation, suggesting that this longer tract may occasionally form a structure that requires Dna2 for correct processing. One possibility is that the helicase activity of Dna2 could serve to unwind secondary structure in the flap, such as that caused by a CTG repeat, which would inhibit cleavage by Rad27 (6).
Since Rad27 activity appears to bypass the need for RNase HI, it has been proposed that the role of RNase HI may be to process 5′ ends that are either annealed to the template or have secondary structure that would inhibit efficient Rad27 cleavage (62). Flaps containing CTG or CAG repeats would be expected to fall into the latter category. A deletion in RNH35 led to a very slight increase in CAG expansions for the longer CAG-70 and -155 tracts and also slightly (but significantly) increased fragility of CAG-155, suggesting that RNase HI may have a role in processing structure-containing flaps in yeast, but another protein (most likely Rad27) can usually substitute for lack of RNase HI activity.
The Pol δ and primase mutations are expected to affect lagging-strand replication, but not processing of the 5′ end of the Okazaki fragment per se. Both mutations showed significant increases in contractions. Since the pol3-t mutation is within a polymerization domain, this enzyme is hypothesized to dissociate from the DNA template more often or exibit slower replication (88). This behavior could leave large portions of the template strand single stranded, which would favor secondary structure formation on the template strand that could lead to contractions. Similarly, inefficient priming in the pri2-1 strain could also lead to longer regions of single-stranded template that would favor contractions. In addition, the pri2-1 allele showed a significant increase in expansions, but only for the CAG-155 tract, which is the same tract length that showed a significant increase in breakage. Perhaps a subtle difference at the 5′ end of the Okazaki fragment (for example, a shorter-than-normal RNA primer) alters the processing of the 5′ flap enough to create a problem in the context of a long CAG tract: for example, by increasing the chance of hairpin formation. Alternatively, the primase defect could inhibit efficient repair of CAG breaks, since primase has been shown to be required for normal levels of DSB repair (34).
The use of a CAG TNR as a potential site of telomere addition is a new observation. It is perhaps relevant that both a rad27Δ and the pri2-1 allele affect telomeres in other assays (21, 64). In a rad27Δ strain grown at 37°C, the template strand for lagging-strand synthesis (G-rich strand) at telomeres has an increased level of single-stranded character, suggesting that the terminal Okazaki fragments may dissociate from the chromosome ends (64). Consistent with this result, we find that CAG tracts (but not unique sequences) show a length-dependent increase in single-stranded character in a rad27Δ strain grown at 37°C and, to a lesser extent, at 30°C (C. H. Freudenreich, unpublished data). On the YAC used, the CAG strand is predicted to be the template for lagging-strand synthesis. Thus, in a rad27Δ strain, breakage in a long CAG/CTG tract might lead to a terminal single-strand CAG tail that could be similar enough to the natural telomerase substrate to act as a site of telomere addition. One can imagine a similar phenomenon in the primase-deficient strain, since deficient primer synthesis on the lagging strand would also be expected to lead to more single-stranded regions on the lagging-strand template (1, 21). These results show that in some cases, breakage of a CAG/CTG TNR in vivo leads to telomere healing and chromosome truncation and suggest another way TNRs could contribute to genomic instability in human cells.
In summary, our data show that mutations that increase CAG expansions also increase CAG fragility and suggest a relationship between the two. One possibility is that each of the mutants that affect triplet repeats increases the probability of secondary structure, such as hairpin formation, but the exact location of that structure (i.e., on the template or nascent strand or within or at the 5′ end of an Okazaki fragment) determines the pathway of repair and thus the outcome. TNR expansions appear more likely to occur because of inefficient Okazaki fragment processing and contractions by inefficient replication of the lagging strand, presumably leading to increased single-stranded regions of the template that could form hairpin structures that would be bypassed by the replication complex. We show that replication mutants can exacerbate triplet repeat fragility and suggest that nicks or breaks that occur within triplet repeats are subject to repair, with the choice of pathway leading to a propensity for either large expansions, small expansions, contractions, or abortive or no repair, resulting in a persistent break detectable in the YAC breakage assay. The pathway of repair could also be influenced by the effect of the mutation itself on various repair pathways. These results have provided insight into how cells usually prevent expansion or fragility and how mutation of some proteins could increase genome instability by exacerbating the problem of unrepaired structures involving long triplet repeats or other at-risk motifs.
Acknowledgments
This work was supported by NIH grant GM63066 to C.H.F. and CA79441 to V.A.Z.
We thank Susan VanReenan for help with fluctuation analyses; Laura Sacco for creation of the RNase H(35) deletion strain; Sandra Schnakenberg, Mark Cooperman, and Alison Lauer for technical assistance; Durwood Marshall and Sara Lewis for help with statistics; Dmitry Gordinen, Michael Resnick, Lee Hartwell, Marco Muzi-Falconi, Paolo Plevani, Martin Budd, and Judith Campbell for reagents; and Brian Lenzmeier and members of the Freudenreich laboratory for helpful comments.
REFERENCES
- 1.Adams Martin, A., I. Dionne, R. J. Wellinger, and C. Holm. 2000. The function of DNA polymerase α at telomeric G tails is important for telomere homeostasis. Mol. Cell. Biol. 20:786-796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anderson, S., and M. L. DePamphilis. 1979. Metabolism of Okazaki fragments during simian virus 40 DNA replication. J. Biol. Chem. 254:11495-11504. [PubMed] [Google Scholar]
- 3.Ayyagari, R., X. V. Gomes, D. A. Gordenin, and P. M. Burgers. 2003. Okazaki fragment maturation in yeast. I. Distribution of functions between FEN1 and DNA2. J. Biol. Chem. 278:1618-1625. [DOI] [PubMed] [Google Scholar]
- 4.Bae, S. H., K. H. Bae, J. A. Kim, and Y. S. Seo. 2001. RPA governs endonuclease switching during processing of Okazaki fragments in eukaryotes. Nature 412:456-461. [DOI] [PubMed] [Google Scholar]
- 5.Bae, S.-H., E. Choi, K.-H. Lee, J. S. Park, S.-H. Lee, and Y.-S. Seo. 1998. Dna2 of Saccharomyces cerevisiae possesses a single-stranded DNA-specific endonuclease activity that is able to act on double-stranded DNA in the presence of ATP. J. Biol. Chem. 273:26880-26890. [DOI] [PubMed] [Google Scholar]
- 6.Bae, S. H., D. W. Kim, J. Kim, J. H. Kim, D. H. Kim, H. D. Kim, H. Y. Kang, and Y. S. Seo. 2002. Coupling of DNA helicase and endonuclease activities of yeast Dna2 facilitates Okazaki fragment processing. J. Biol. Chem. 277:26632-26641. [DOI] [PubMed] [Google Scholar]
- 7.Bae, S. H., and Y. S. Seo. 2000. Characterization of the enzymatic properties of the yeast Dna2 helicase/endonuclease suggests a new model for Okazaki fragment processing. J. Biol. Chem. 275:38022-38031. [DOI] [PubMed] [Google Scholar]
- 8.Balakumaran, B. S., C. H. Freudenreich, and V. A. Zakian. 2000. CGG/CCG repeats exhibit orientation-dependent instability and orientation-independent fragility in Saccharomyces cerevisiae. Hum. Mol. Genet. 9:93-100. [DOI] [PubMed] [Google Scholar]
- 9.Bambara, R. A., R. S. Murante, and L. A. Henricksen. 1997. Enzymes and reactions at the eukaryotic DNA replication fork. J. Biol. Chem. 272:4647-4650. [DOI] [PubMed] [Google Scholar]
- 10.Baudin, A., O. Ozier-Kalogeropoulos, A. Denouel, F. Lacroute, and C. Cullen. 1993. A simple and efficient method for direct gene deletion in Saccharomyces cerevisiae. Nucleic Acids Res. 21:3329-3330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bielinsky, A. K., and S. A. Gerbi. 1998. Discrete start sites for DNA synthesis in the yeast ARS1 origin. Science 279:95-98. [DOI] [PubMed] [Google Scholar]
- 12.Budd, M. E., and J. L. Campbell. 1995. A yeast gene required for DNA replication encodes a protein with homology to DNA helicases. Proc. Natl. Acad. Sci. USA 92:7642-7646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Budd, M. E., and J. L. Campbell. 1997. A yeast replicative helicase, Dna2 helicase, interacts with yeast FEN-1 nuclease in carrying out its essential function. Mol. Cell. Biol. 17:2136-2142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Budd, M. E., W. Choe, and J. L. Campbell. 2000. The nuclease activity of the yeast DNA2 protein, which is related to the RecB-like nucleases, is essential in vivo. J. Biol. Chem. 275:16518-16529. [DOI] [PubMed] [Google Scholar]
- 15.Chen, C., and R. Kolodner. 1999. Gross chromosomal rearrangements in Saccharomyces cerevisiae replication and recombination defective mutants. Nat. Genet. 23:81-85. [DOI] [PubMed] [Google Scholar]
- 16.Cleary, J. D., K. Nichol, Y. H. Wang, and C. E. Pearson. 2002. Evidence of cis-acting factors in replication-mediated trinucleotide repeat instability in primate cells. Nat. Genet. 31:37-46. [DOI] [PubMed] [Google Scholar]
- 17.Cleary, J. D., and C. E. Pearson. The contribution of cis-elements to disease-associated repeat instability. Cytogenet. Genome Res., in press. [DOI] [PubMed]
- 18.Cohen, H., D. D. Sears, D. Zenvirth, P. Hieter, and G. Simchen. 1999. Increased instability of human CTG repeat tracts on yeast artificial chromosomes during gametogenesis. Mol. Cell. Biol. 19:4153-4158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cummings, C. J., and H. Y. Zoghbi. 2000. Fourteen and counting: unraveling trinucleotide repeat diseases. Hum. Mol. Genet. 9:909-916. [DOI] [PubMed] [Google Scholar]
- 20.Debrauwere, H., S. Loeillet, W. Lin, J. Lopes, and A. Nicolas. 2001. Links between replication and recombination in Saccharomyces cerevisiae: a hypersensitive requirement for homologous recombination in the absence of Rad27 activity. Proc. Natl. Acad. Sci. USA 98:8263-8269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Diede, S. J., and D. E. Gottschling. 1999. Telomerase-mediated telomere addition in vivo requires DNA primase and DNA polymerases alpha and delta. Cell 99:723-733. [DOI] [PubMed] [Google Scholar]
- 22.Dutcher, S. K. 1981. Internuclear transfer of genetic information in kar1/KAR1 heterokaryons in Saccharomyces cerevisiae. Mol. Cell. Biol. 1:245-253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Francesconi, S., M. P. Longhese, A. Piseri, C. Santocanale, G. Lucchini, and P. Plevani. 1991. Mutations in conserved yeast DNA primase domains impair DNA replication in vivo. Proc. Natl. Acad. Sci. USA 88:3877-3881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Frank, P., C. Craunshofer-Reiter, and U. Wintersberger. 1998. Yeast RNase H(35) is the counterpart of the mammalian RNase HI, and is evolutionarily related to prokaryotic RNase HII. FEBS Lett. 421:23-26. [DOI] [PubMed] [Google Scholar]
- 25.Freudenreich, C. H., S. M. Kantrow, and V. Zakian. 1998. Expansion and length-dependent fragility of CTG repeats in yeast. Science 279:853-856. [DOI] [PubMed] [Google Scholar]
- 26.Freudenreich, C. H., J. B. Stavenhagen, and V. A. Zakian. 1997. Stability of a CTG/CAG trinucleotide repeat in yeast is dependent on its orientation in the genome. Mol. Cell. Biol. 17:2090-2098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gacy, A. M., G. Goellner, N. Juranic, S. Macura, and C. T. McMurray. 1995. Trinucleotide repeats that expand in human disease form hairpin structures in vitro. Cell 81:533-540. [DOI] [PubMed] [Google Scholar]
- 28.Gary, R., M. S. Park, J. P. Nolan, H. L. Cornelius, O. G. Kozyreva, H. T. Tran, K. S. Lobachev, M. A. Resnick, and D. A. Gordenin. 1999. A novel role in DNA metabolism for the binding of Fen1/Rad27 to PCNA and implications for genetic risk. Mol. Cell. Biol. 19:5373-5382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gordenin, D. A., T. A. Kunkel, and M. A. Resnick. 1997. Repeat expansion—all in a flap? Nat. Genet. 16:116-118. [DOI] [PubMed] [Google Scholar]
- 30.Hartenstine, M. J., M. F. Goodman, and J. Petruska. 2002. Weak strand displacement activity enables human DNA polymerase beta to expand CAG/CTG triplet repeats at strand breaks. J. Biol. Chem. 277:41379-41389. [DOI] [PubMed] [Google Scholar]
- 31.Hartwell, L. H., R. K. Mortimer, J. Culotti, and M. Culotti. 1973. Genetic control of the cell division cycle in yeast: genetic analysis of cdc mutants. Genetics 74:267-286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Henricksen, L. A., S. Tom, Y. Liu, and R. A. Bambara. 2000. Inhibition of flap endonuclease 1 by flap secondary structure and relevance to repeat sequence expansion. J. Biol. Chem. 275:16420-16427. [DOI] [PubMed] [Google Scholar]
- 33.Henricksen, L. A., J. Veeraraghavan, D. R. Chafin, and R. A. Bambara. 2002. DNA ligase I competes with FEN1 to expand repetitive DNA sequences in vitro. J. Biol. Chem. 277:22361-22369. [DOI] [PubMed] [Google Scholar]
- 34.Holmes, A. M., and J. E. Haber. 1999. Double-strand break repair in yeast requires both leading and lagging strand DNA polymerases. Cell 96:415-424. [DOI] [PubMed] [Google Scholar]
- 35.Hosfield, D. J., C. D. Mol, B. Shen, and J. A. Tainer. 1998. Structure of the DNA repair and replication endonuclease and exonuclease FEN-1: coupling DNA and PCNA binding to FEN-1 activity. Cell 95:135-146. [DOI] [PubMed] [Google Scholar]
- 36.Ireland, M. J., S. S. Reinke, and D. M. Livingston. 2000. The impact of lagging strand replication mutations on the stability of CAG repeat tracts in yeast. Genetics 155:1657-1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jakupciak, J. P., and R. D. Wells. 1999. Genetic instabilities in (CTG·CAG) repeats occur by recombination. J. Biol. Chem. 274:23468-23479. [DOI] [PubMed] [Google Scholar]
- 38.Jankowski, C., and D. K. Nag. 2002. Most meiotic CAG repeat tract-length alterations in yeast are SPO11 dependent. Mol. Genet. Genomics 267:64-70. [DOI] [PubMed] [Google Scholar]
- 39.Jankowski, C., F. Nasar, and D. K. Nag. 2000. Meiotic instability of CAG repeat tracts occurs by double-strand break repair in yeast. Proc. Natl. Acad. Sci. USA 97:2134-2139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Johnson, R., G. Kovvali, L. Prakash, and S. Prakash. 1998. Role of yeast Rth1 nuclease and its homologs in mutation avoidance, DNA repair, and DNA replication. Curr. Genet. 34:21-29. [DOI] [PubMed] [Google Scholar]
- 41.Johnson, R. E., G. K. Kowali, L. Prakash, and S. Prakash. 1995. Requirement of the yeast RTH1 5′ to 3′ exonuclease for the stability of simple repetitive DNA. Science 269:238-240. [DOI] [PubMed] [Google Scholar]
- 42.Kang, S., A. Jaworski, K. Ohshima, and R. D. Wells. 1995. Expansion and deletion of CTG repeats from human disease genes are determined by the direction of replication in E. coli. Nat. Genet. 10:213-218. [DOI] [PubMed] [Google Scholar]
- 43.Kao, H.-I., L. A. Henricksen, Y. Liu, and R. A. Bambara. 2002. Cleavage specificity of Saccharomyces cerevisiae Flap endonuclease 1 suggests a double-flap structure as the cellular substrate. J. Biol. Chem. 277:14379-14389. [DOI] [PubMed] [Google Scholar]
- 44.Kim, K., S. Biade, and Y. Matsumoto. 1998. Involvement of flap endonuclease 1 in base excision DNA repair. J. Biol. Chem. 273:8842-8848. [DOI] [PubMed] [Google Scholar]
- 45.Klungland, A., and T. Lindahl. 1997. Second pathway for completion of human DNA base excision-repair: reconstitution with purified proteins and requirement for DNase IV (FEN1). EMBO J. 16:3341-3348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kokoska, R. J., L. Stefanovic, H. T. Tran, M. A. Resnick, D. A. Gordenin, and T. D. Petes. 1998. Destabilization of yeast micro- and minisatellite DNA sequences by mutations affecting a nuclease involved in Okazaki fragment processing (rad27) and DNA polymerase δ (pol3-t). Mol. Cell. Biol. 18:2779-2788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kovtun, I., and C. McMurray. 2001. Trinucleotide expansion in haploid germ cells by gap repair. Nat. Genet. 27:407-411. [DOI] [PubMed] [Google Scholar]
- 48.Kovtun, I. V., G. Goellner, and C. T. McMurray. 2001. Structural features of trinucleotide repeats associated with DNA expansion. Biochem. Cell Biol. 79:325-336. [PubMed] [Google Scholar]
- 49.Lea, D. E., and C. A. Coulson. 1949. The distribution of the numbers of mutants in bacterial populations. J. Genet. 49:264-285. [DOI] [PubMed] [Google Scholar]
- 50.Lee, K. H., D. W. Kim, S. H. Bae, J. A. Kim, G. H. Ryu, Y. N. Kwon, K. A. Kim, H. S. Koo, and Y. S. Seo. 2000. The endonuclease activity of the yeast Dna2 enzyme is essential in vivo. Nucleic Acids Res. 28:2873-2881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lenzmeier, B. A., and C. H. Freudenreich. Trinucleotide repeat instability: a hairpin curve at the crossroads of replication, recombination, and repair. Cytogenet. Genome Res., in press. [DOI] [PubMed]
- 52.Liu, Y., and R. A. Bambara. 2003. Analysis of human flap endonuclease 1 mutants reveals a mechanism to prevent triplet repeat expansion. J. Biol. Chem. 278:13728-13739. [DOI] [PubMed] [Google Scholar]
- 53.Lopes, J., H. Debrauwere, J. Buard, and A. Nicolas. 2002. Instability of the human minisatellite CEB1 in rad27Delta and dna2-1 replication-deficient yeast cells. EMBO J. 21:3201-3211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lyons-Darden, T., and M. D. Topal. 1999. Abasic sites induce triplet-repeat expansion during DNA replication in vitro. J. Biol. Chem. 274:25975-25978. [DOI] [PubMed] [Google Scholar]
- 55.Maleki, S., H. Cederberg, and U. Rannug. 2002. The human minisatellites MS1, MS32, MS205 and CEB1 integrated into the yeast genome exhibit different degrees of mitotic instability but are all stabilised by RAD27. Curr. Genet. 41:333-341. [DOI] [PubMed] [Google Scholar]
- 56.Manley, K., T. L. Shirley, L. Flaherty, and A. Messer. 1999. Msh2 deficiency prevents in vivo somatic instability of the CAG repeat in Huntington disease transgenic mice. Nat. Genet. 23:471-473. [DOI] [PubMed] [Google Scholar]
- 57.Maurer, D. J., B. L. O'Callaghan, and D. M. Livingston. 1996. Orientation dependence of trinucleotide CAG repeat instability in Saccharomyces cerevisiae. Mol. Cell. Biol. 16:6617-6622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Michel, B. 2000. Replication fork arrest and DNA recombination. Trends Biochem. Sci. 25:173-178. [DOI] [PubMed] [Google Scholar]
- 59.Michel, B., S. D. Erlich, and M. Uzest. 1997. DNA double-strand breaks caused by replication arrest. EMBO J. 16:430-438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Miret, J. J., L. Pessoa-Brandão, and R. S. Lahue. 1997. Instability of CAG and CTG trinucleotide repeats in Saccharomyces cerevisiae. Mol. Cell. Biol. 17:3382-3387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mitas, M. 1997. Trinucleotide repeats associated with human disease. Nucleic Acids Res. 25:2245-2253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Murante, R. S., L. A. Henricksen, and R. A. Bambara. 1998. Junction ribonuclease: an activity in Okazaki fragment processing. Proc. Natl. Acad. Sci. USA 95:2244-2249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Nelson, D. L. 1995. The fragile X syndromes. Cell Biol. 6:5-11. [DOI] [PubMed] [Google Scholar]
- 64.Parenteau, J., and R. J. Wellinger. 1999. Accumulation of single-stranded DNA and destabilization of telomeric repeats in yeast mutant strains carrying a deletion of RAD27. Mol. Cell. Biol. 19:4143-4152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Pearson, C. E., and R. R. Sinden. 1998. Trinucleotide repeat DNA structures: dynamic mutations from dynamic DNA. Curr. Opin. Struct. Biol. 8:321-330. [DOI] [PubMed] [Google Scholar]
- 66.Pelletier, R., M. M. Krasilnikova, G. M. Samadashwily, R. Lahue, and S. M. Mirkin. 2003. Replication and expansion of trinucleotide repeats in yeast. Mol. Cell. Biol. 23:1349-1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Pluta, A. F., and V. A. Zakian. 1989. Recombination occurs during telomere formation in yeast. Nature 337:429-433. [DOI] [PubMed] [Google Scholar]
- 68.Qiu, J., Y. Qian, P. Frank, U. Wintersberger, and B. Shen. 1999. Saccharomyces cerevisiae RNase H(35) functions in RNA primer removal during lagging-strand DNA synthesis, most efficiently in cooperation with Rad27 nuclease. Mol. Cell. Biol. 19:8361-8371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Reagan, M. S., C. Pittenger, W. Siede, and E. C. Friedberg. 1995. Characterization of a mutant strain of Saccharomyces cerevisiae with a deletion of the RAD27 gene, a structural homolog of the RAD2 nucleotide excision repair gene. J. Bacteriol. 177:364-371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Richard, G.-F., B. Dujon, and J. E. Haber. 1999. Double-strand break repair can lead to high frequencies of deletions within short CAG/CTG trinucleotide repeats. Mol. Gen. Genet. 261:871-882. [DOI] [PubMed] [Google Scholar]
- 71.Richard, G. F., G. M. Goellner, C. T. McMurray, and J. E. Haber. 2000. Recombination-induced CAG trinucleotide repeat expansions in yeast involve the MRE11-RAD50-XRS2 complex. EMBO J. 19:2381-2390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Richard, G. F., and F. Paques. 2000. Mini- and microsatellite expansions: the recombination connection. EMBO Rep. 1:122-126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Richards, R. I. 2001. Dynamic mutations: a decade of unstable expanded repeats in human genetic disease. Hum. Mol. Genet. 10:2187-2194. [DOI] [PubMed] [Google Scholar]
- 74.Rothstein, R., B. Michel, and S. Gangloff. 2000. Replication fork pausing and recombination or “gimme a break.” Genes Dev. 14:1-10. [PubMed] [Google Scholar]
- 75.Samadashwily, G. M., G. Raca, and S. M. Mirkin. 1997. Trinucleotide repeats affect DNA replication in vivo. Nat. Genet. 17:298-304. [DOI] [PubMed] [Google Scholar]
- 76.Sarkar, P. S., H.-C. Chang, B. Boudi, and S. Reddy. 1998. CTG repeats show bimodal amplification in E. coli. Cell 95:531-540. [DOI] [PubMed] [Google Scholar]
- 77.Schulz, V. P., and V. A. Zakian. 1994. The Saccharomyces PIF1 DNA helicase inhibits telomere elongation and de novo telomere formation. Cell 76:145-155. [DOI] [PubMed] [Google Scholar]
- 78.Schweitzer, J. K., and D. M. Livingston. 1999. The effect of DNA replication mutations on CAG tract stability in yeast. Genetics 152:953-963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Schweitzer, J. K., and D. M. Livingston. 1998. Expansions of CAG repeat tracts are frequent in a yeast mutant defective in Okazaki fragment maturation. Hum. Mol. Genet. 7:69-74. [DOI] [PubMed] [Google Scholar]
- 80.Schweitzer, J. K., S. S. Reinke, and D. M. Livingston. 2001. Meiotic alterations in CAG repeat tracts. Genetics 159:1861-1865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Shibata, Y., and T. Nakamura. 2002. Defective flap endonuclease 1 activity in mammalian cells is associated with impaired DNA repair and prolonged S phase delay. J. Biol. Chem. 277:746-754. [DOI] [PubMed] [Google Scholar]
- 82.Sinden, R. R. 1999. Human Genetics '99: trinucleotide repeats. Biological implications of the DNA structures associated with disease-causing triplet repeats. Am. J. Hum. Genet. 64:346-353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Sommers, C. H., E. J. Miller, B. Dujon, S. Prakash, and L. Prakash. 1995. Conditional lethality of null mutations in RTH1 that encodes the yeast counterpart of a mammalian 5′- to 3′-exonuclease required for lagging strand synthesis in reconstituted systems. J. Biol. Chem. 270:4193-4196. [DOI] [PubMed] [Google Scholar]
- 84.Spiro, C., R. Pelletier, M. L. Rolfsmeir, M. J. Dixon, R. S. Lahue, G. Gupta, M. S. Park, X. Chen, S. V. Mariapp, and C. T. McMurray. 1999. Inhibition of FEN-1 processing by DNA secondary structure at trinucleotide repeats. Mol. Cell 4:1079-1085. [DOI] [PubMed] [Google Scholar]
- 85.Sutherland, G. R., E. Baker, and R. I. Richards. 1998. Fragile sites still breaking. Trends Genet. 14:501-506. [DOI] [PubMed] [Google Scholar]
- 86.Symington, L. S. 1998. Homologous recombination is required for the viability of rad27 mutants. Nucleic Acids Res. 26:5589-5595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Tishkoff, D. X., N. Filosi, G. A. Gaida, and R. D. Kolodner. 1997. A novel mutation avoidance mechanism dependent on S. cerevisiae RAD27 is distinct from DNA mismatch repair. Cell 88:253-263. [DOI] [PubMed] [Google Scholar]
- 88.Tran, H. T., N. P. Degtyareva, D. A. Gordenin, and M. A. Resnick. 1999. Genetic factors affecting the impact of DNA polymerase δ activity on mutation avoidance in yeast. Genetics 152:47-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Tran, H. T., N. P. Degtyareva, N. N. Koloteva, A. Sugino, H. Masumoto, D. A. Gordenin, and M. A. Resnick. 1995. Replication slippage between distant short repeats in Saccharomyces cerevisiae depends on the direction of replication and the RAD50 and RAD52 genes. Mol. Cell. Biol. 15:5607-5617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.van den Broek, W. J., M. R. Nelen, D. G. Wansink, M. M. Coerwinkel, H. te Riele, P. J. Groenen, and B. Wieringa. 2002. Somatic expansion behaviour of the (CTG)n repeat in myotonic dystrophy knock-in mice is differentially affected by Msh3 and Msh6 mismatch-repair proteins. Hum. Mol. Genet. 11:191-198. [DOI] [PubMed] [Google Scholar]
- 91.Wu, X., E. Braithwaite, and Z. Wang. 1999. DNA ligation during excision repair in yeast cell-free extracts is specifically catalyzed by the CDC9 gene product. Biochemistry 38:2628-2635. [DOI] [PubMed] [Google Scholar]
- 92.Wu, X., and Z. Wang. 1999. Relationships between yeast Rad27 and Apn1 in response to apurinic/apyrimidinic (AP) sites in DNA. Nucleic Acids Res. 27:956-962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Wu, X., T. E. Wilson, and M. R. Lieber. 1999. A role for FEN-1 in nonhomologous DNA end joining: the order of strand annealing and nucleolytic processing events. Proc. Natl. Acad. Sci. USA 96:1303-1308. [DOI] [PMC free article] [PubMed] [Google Scholar]