Abstract
PU.1 and GATA-1 are two hematopoietic specific transcription factors that play key roles in development of the myeloid and erythroid lineages, respectively. The two proteins bind to one another and inhibit each other's function in transcriptional activation and promotion of their respective differentiation programs. This mutual antagonism may be an important aspect of lineage commitment decisions. PU.1 can also act as an oncoprotein since deregulated expression of PU.1 in erythroid precursors causes erythroleukemias in mice. Studies of cultured mouse erythroleukemia cell lines indicate that one aspect of PU.1 function in erythroleukemogenesis is its ability to block erythroid differentiation by repressing GATA-1 (N. Rekhtman, F. Radparvar, T. Evans, and A. I. Skoultchi, Genes Dev. 13:1398-1411, 1999). We have investigated the mechanism of PU.1-mediated repression of GATA-1. We report here that PU.1 binds to GATA-1 on DNA. We localized the repression activity of PU.1 to a small acidic N-terminal domain that interacts with the C pocket of pRB, a well-known transcriptional corepressor. Repression of GATA-1 by PU.1 requires pRB, and pRB colocalizes with PU.1 and GATA-1 at repressed GATA-1 target genes. PU.1 and pRB also cooperate to block erythroid differentiation. Our results suggest that one of the mechanisms by which PU.1 antagonizes GATA-1 is by binding to it at GATA-1 target genes and tethering to these sites a corepressor that blocks transcriptional activity and thereby erythroid differentiation.
Recently, much progress has been made in identifying the transcription factors needed for development of specific hematopoietic lineages and, to some extent, their hierarchical relationships (44). There is evidence that these factors act in a combinatorial manner, in some cases through direct protein-protein interactions (7, 54). Such interactions may either stimulate or inhibit transcription factor activity. Accumulating evidence indicates that gene-specific repression of transcription is as important as transcriptional activation in normal development (9). Thus, extinction of one transcriptional program may be as important as activation of an alternative program for correct developmental decisions. However, inappropriate or untimely transcriptional repression in immature cells is also thought to be the basis for the block to differentiation present in several types of hematologic malignancies (32, 34, 58).
PU.1 and GATA-1 are two hematopoiesis-specific transcription factors that physically interact and can antagonize each other's transcriptional activity and ability to promote their respective differentiation programs (39, 43, 50, 71). PU.1 is an Ets family member that is required for the development of myeloid cells and B cells (13, 55). Enforced expression of PU.1 blocks erythroid differentiation in cultured cells and in embryos (11, 45, 46, 50, 53, 66), and activation of PU.1 expression through provirus integration in immature erythroid precursor cells is a critical event in generation of Friend virus-induced erythroleukemias in mice (3, 41, 42). GATA-1 is a zinc finger protein that is essential for red blood cell development (7). Expression of GATA-1 in avian multipotential precursor cells or in an early myeloid cell line, 416B, blocks myeloid differentiation (31, 61). Expression of GATA-1 mRNA has been seen in human myeloid leukemic cells (15). Taken together, these data indicate that the mutual antagonism exhibited by PU.1 and GATA-1 may be important in processes controlling normal hematopoietic development, as well as during malignant transformation in leukemia.
Both PU.1 and GATA-1 can act as transcriptional activators. Numerous genes that are direct targets of their transcriptional stimulatory activity have been identified in myeloid and lymphoid cells and erythroid cells, respectively. These genes contain sequence-specific binding sites for the respective factor. Both proteins contain regions that are essential for DNA binding: the Ets region in PU.1 and the zinc finger region in GATA-1 (28, 38). These regions are also involved in the PU.1-GATA-1 interaction (50). Both proteins also engage in specific interactions with other proteins that enhance their transcriptional activity, including PU.1 with c-Jun in myeloid cells (2) and with PU.1 interacting partner (Pip) in B cells (12) and GATA-1 with Friend of GATA-1 (FOG-1) in erythroid cells (60). Both proteins also interact with the ubiquitous coactivator CBP (4, 67). Thus, direct physical interaction of PU.1 and GATA-1 could mediate inhibition of their transcriptional activity at several different levels. Both PU.1 and GATA-1 have been reported to interact with components of transcriptional repression complexes (see Discussion). The formation of a ternary complex of PU.1, GATA-1, and one of these components could also lead to inhibition of transcriptional activity at PU.1 or GATA-1 target genes.
GATA-1 has been reported to inhibit PU.1 function by interfering with PU.1 binding to c-Jun, an important PU.1 cofactor in myeloid cells (71). On the other hand, PU.1 does not interfere with in vitro binding of GATA-1 to its cofactors FOG-1 or CBP (49) and complexes of GATA-1 with FOG-1 and CBP have been detected in mouse erythroleukemia (MEL) cells (4, 60) that contain high levels of PU.1, which complexes with GATA-1 in the cells and blocks their ability to differentiate (50). GATA-1 has been reported not to interfere with PU.1 binding to DNA (71), but others have reported that it does inhibit PU.1 DNA binding (39). There is also conflicting evidence regarding a role for inhibition of DNA binding in PU.1-mediated repression of GATA-1. PU.1 has been reported to inhibit GATA-1 DNA binding in vitro (39, 72). A decrease in in vitro GATA-1 DNA-binding activity also was reported in extracts of MEL cells overexpressing PU.1 (65). However, others did not observe a decrease in GATA-1 DNA binding in such cells (20), and there are several reports indicating that this activity is not increased in MEL cells when PU.1 levels decline as the cells undergo differentiation (1, 10, 20). Moreover, in vivo footprinting experiments have shown that GATA-1 binding sites in several erythroid cell-specific promoters are occupied in undifferentiated MEL cells that contain high levels of PU.1 and that occupancy does not change as PU.1 levels decline during differentiation (48, 56). Chromatin immunoprecipitation (ChIP) experiments also show that GATA-1 is present at erythroid-specific promoters in MEL cells (68). Thus, although PU.1 may be capable of inhibiting GATA-1 DNA binding in vitro, the mechanism by which it represses GATA-1 transcriptional activity and function in erythroid differentiation in vivo remains to be defined.
We reported previously that, whereas the C-terminal Ets homology region of PU.1 is necessary and sufficient for PU.1 binding to GATA-1 both in vitro and in vivo, the N-terminal region is also needed for repression of GATA-1, as well as for its ability to inhibit erythroid differentiation by PU.1. Based on these observations, we suggested that PU.1 might require a cofactor to help mediate repression of GATA-1 (50). We report here that an established corepressor, pRB, binds to a small acidic domain in the N-terminal region of PU.1 and that it cooperates with PU.1 in mediating repression of GATA-1 and inhibition of erythroid differentiation.
MATERIALS AND METHODS
ChIP.
A total of 107 U2OS cells were plated in 15-cm plates 1 day prior to transfection. Cells were transfected by the Lipofectamine Plus method (Gibco-BRL), harvested after 36 h, and treated with 1% formaldehyde for 30 min at room temperature with gentle shaking. Cell extract preparation and immunoprecipitation were performed as described previously (5). The antibodies used included 8 μl of anti-GATA-1 antibody (N6; Santa Cruz) plus 2 μl of anti-rat antibody (Sigma), 15 μl of anti-PU.1 antibody (T21; Santa Cruz), 15 μl of anti-hemagglutinin (HA) antibody (SC-805; Santa Cruz), 15 μl of cyclin E antibody (M-20; Santa Cruz), 5 μl of anti-pRB antibody (C-15; Santa Cruz), or 2.5 μl of anti-rabbit immunoglobulin G antibody (SC-2027; Santa Cruz). Five to ten percent of the DNA extracted from the immunoprecipitates was used for PCRs. PCR was performed at 55°C for 30 cycles with T3 primer and an αD3 primer (CTCTAGAGGATCCCCTGG), yielding a 180-bp DNA product, or for 25 cycles with ATTCCTGCAGGGCCGACCTC and TTCTGCAGGTACCGAGCTCC, which yielded a 110-bp DNA product.
For endogenous ChIP of MEL cell chromatin (clone DS19), preparation of cross-linked chromatin and immunoprecipitation were performed essentially as described elsewhere (57), with the following modifications. Nuclei from 108 MEL cells were resuspended in 4 ml and sonicated (40%, 16 cycles of 30 s, in a dry ice-ethanol cooling bath) with a Branson Sonic Dismembrator model 500 equipped with a microtip to yield 200- to 400-bp DNA fragments. Immunoprecipitations were performed by incubating sheared chromatin (precleared with beads for 2 h) from 2.5 × 106 cells overnight at 4°C with 1 μg of antibody bound to 8 μl of a 1:1 mixture of protein G- and protein A-Sepharose beads. Antibodies used were as described above and also included anti-Rb (G3-245; Pharmingen) and a control antibody, anti-β-tubulin (ab6161; Novus Biologicals, Inc.). During binding of the antibody to the beads, the samples were also incubated with sonicated salmon sperm DNA (40 μg/immunoprecipitation) and bovine serum albumin (400 μg/immunoprecipitation). The amounts of each specific DNA fragment in immunoprecipitates were determined relative to the amount of the fragment in input DNA by measuring the difference (dx) in the CT values of the immunoprecipitated sample and the input DNA. The fold enrichment (FE) was calculated as FE = 2−dCT where dCT = dG6PD − dGATA-1. Use of an antibody to an irrelevant antigen (e.g., β-tubulin; see Fig. 3B) or an isotype control antibody (data not shown) gave FE values of approximately 1. Results similar to those shown in Fig. 3B were also obtained by using either regions of the GAPDH (glyceraldehyde-3-phosphate dehydrogenase) or MyoD promoters as non-GATA-1-binding control sequences instead of the glucose 6-phosphate dehydrogenase (G6PD) promoter. The dx values for these sites with specific antibodies (against GATA-1, PU.1 or pRB) compared to control antibodies were similar, indicating that these sites were indeed not occupied by GATA-1, PU.1, or pRB.
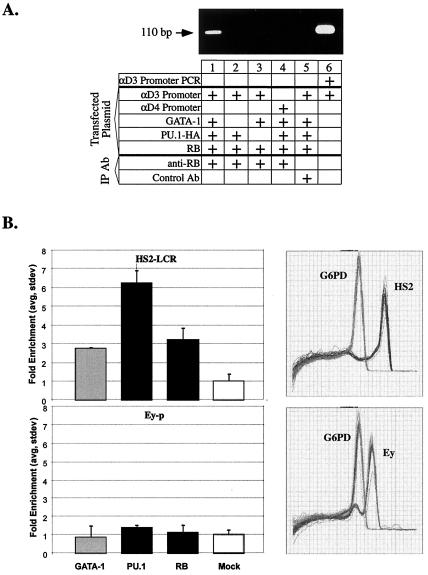
FIG. 3.
PU.1 and pRB colocalize with GATA-1 at GATA-1 binding sites in chromatin. (A) U2OS cells were transfected as described in the legend to Fig. 1A, and ChIP was carried out as described in Materials and Methods with anti-pRB antibody or an anti-rabbit immunoglobulin G antibody that has the same isotype (Control Ab). PCR assays were performed as described in Materials and Methods with specific primers that differ from the primers used in Fig. 1A; here the presence of the 110-bp fragment indicates the presence of the αD3 promoter region in the immunoprecipitate. Lane 6 shows PCR amplification of αD3-Luc plasmid DNA with the primers. (B) ChIP was performed on cross-linked chromatin from MEL cells as described in Materials and Methods with antibodies to GATA-1, PU.1, and pRB (RB) and with β-tubulin (Mock). The amounts of specific promoter DNA fragments relative to G6PD DNA in duplicate immunoprecipitates were quantitated by real-time PCRs, and the degree of enrichment of the promoter fragments was calculated. Error bars indicate the standard deviation. Panels on the right show the dissociation curves of the set of amplified DNA fragments for HS2 (Tm = 85°C), G6PD (Tm = 78.5°C), and Ey (Tm = 82°C).
In vitro protein interaction studies.
Preparation of glutathione S-transferase (GST)-fusion proteins and binding assay conditions were as described previously (50). [35S]methionine-labeled proteins were prepared by coupled transcription- translation reactions in reticulocyte lysates (Promega). Plasmids for GST-pRB and pRB mutant proteins were kindly provided by W. G. Kaelin (25) and A. S. Yee (22). The plasmids used for in vitro transcription-translation reactions included pGEM-HDAC1, pCMV-NCoR/SMRT, pCDNA-mSin3a, pCDNA-mSin3b, pCDNA-RbAp48, and pCDNA-SAP18 (all kindly provided by L. Alland); pGEM-PU.1 (46); PU.1-ΔPEST (50); pBKS-PU.1-ΔDEQ (identical to pBSK-PU.1-ΔTAD in reference 50); and pBSK-PU.1-ΔDE, kindly provided by M. J. Klemsz (mutant NP in reference 27). pGEM-PU.1-ΔQ was constructed by digesting pGEM-PU.1 with PstI and NcoI enzymes, followed by blunt-end self-ligation. pBSK-pRB was generated by subcloning a BamHI fragment of pRB, derived from pLitmus-pRB (generously provided by L. Zhu) into BamHI-digested pBSK. pBSK-pRB-C, containing the 550 C-terminal nucleotides of pRB, was provided by T. Evans.
In vivo protein interaction studies.
Cells were grown as described previously (46) and transfected by the Lipofectamine Plus or calcium phosphate precipitation methods. Whole-cell extracts were prepared 36 h after transfection as described previously (50). Expression vectors included pCMV-pRB, encoding the large pocket region of pRB (amino acids 379 to 928), kindly provided by L. Zhu, pCMV-E2F1, a kind gift from R. G. Pestell, and pEBB-PU.1 (46). For immunoprecipitation of pRB, cell extracts were incubated with 50 μl of protein A-Sepharose beads (Pharmacia) and either anti-human pRB antibody XZ55 (15 μl; Pharmingen) or anti-human pRB antibody G3-245 (15 μl; Pharmingen). For immunoprecipitation of PU.1, 50 μl of protein A-Sepharose beads and 15 μl of anti-PU.1 antibody (T21, Santa Cruz) were used. Then, 1 mg of MEL cell extract or 400 μg of extracts from transiently transfected cells was used for immunoprecipitation.
Reporter assays.
Cell lines were cultured as described above, and 1 day prior to transfection 3 × 104 cells were plated in each well of 24-well plates. Cells were transfected with 220 ng of DNA, 3 μl of Plus reagent, and 2 μl of Lipofectamine reagent, as recommended by the manufacturer (Gibco-BRL). The total amount of DNA was maintained at 220 ng/well by adding the appropriate amount of pBluescript vector DNA. Luciferase production was measured after 36 h with the Promega Luciferase Assay system. Trichostatin A (TSA; Waco BioProducts) was added 24 h after transfection, and the luciferase activity was determined 24 h after TSA addition. Plasmids utilized in reporter assays included pCMV-RB (C706F), encoding the large pocket of pRB with a C-to-F substitution in residue 706, kindly provided by L. Zhu. RB-C-pocket and GAL4-E2F1 expression vectors were kindly provided by T. Evans and R. G. Pestell, respectively. GATA-GAL4(DBD), a gift from T. Evans, contains the GAL4 DNA-binding domain (residues 1 to 147) replacing the C finger of chicken GATA-1 in pHY41-2 vector. Reporter plasmids included αD3-LUC, αD4-LUC (17), and (UAS)5E1BTATA-LUC (62). Values in reporter assays represent the average of at least two independent transfections. Error bars represent the standard error of the mean. All experiments were performed at least twice.
Preparation and analysis of MEL cell stable transfectants.
MEL cells (clone DS19) were cultured and transfected with the lipofectin reagent (Gibco-BRL) as described previously (50). Cells were plated at 103 cells/well in 96-well plates and selection with 5 μg of puromycin/ml was initiated after 24 h. Puromycin-resistant clones were expanded, and cell extracts were analyzed by immunoblotting. Benzidine staining was performed as described previously (46). PU.1-ΔDEQ-HA, PU.1-ΔPEST-HA, PU.1-ΔDE-HA, and PU.1-ΔQ-HA were prepared by substitution of the BamHI-KpnI fragment of pEBB-PU.1-HA-puro (50) with mutated inserts derived from the respective constructs in pBKS. Expression of each construct was verified by immunoblot analysis of transfected 293 cells prior to generating stable MEL cell lines. The sequences encoding the pRB-PU.1ΔN fusion protein were created as follows: pRB residues 1932 to 2790 were amplified by PCR with 5′ primer (5′-ATCTACCTCTCTTTCACTGTT-3′) and 3′ primer (5′-ACTGCCATGGTCTCTTCCTTGTTTGAGGTATC-3′) containing a novel NcoI site encoded in a 5′ “overhang” (underlined) prior to the termination codon. The PCR-generated fragment was digested with NheI and NcoI and ligated with the BamHI-NheI fragment of pRB, derived from pBSK-RB, and NcoI-BamHI-digested pBSK-PU.1 vector (pBSK-pRB-PU.1ΔN). The pEBB-pRB-PU.1ΔN expression construct was created by subcloning a BamHI-NotI fragment derived from pBSK-pRB-PU.1ΔN into the pEBB-puro vector digested with BamHI and NotI. The expression vector encoding the fusion protein, pRB-PU.1ΔN-ΔEts, was created by digesting the pEBB-pRB-PU.1ΔN vector with KpnI and NotI, followed by blunt-end self-ligation.
Immunoblotting.
Western blotting was performed as described previously (50). Primary antibodies included rat monoclonal anti-GATA-1 antibody N6 (1:500; Santa Cruz); rabbit polyclonal anti-PU.1 antibody T21 (1:300; Santa Cruz); mouse monoclonal anti-HA antibody 12CA5 (1:3500; Boehringer Mannheim); anti-human pRB mouse monoclonal antibody XZ55 (1:500; Pharmingen); anti-human pRB mouse monoclonal antibody XZ91 (1:500; Pharmingen); anti-human pRB mouse monoclonal antibody, which is cross-reactive with mouse pRB, G3-245 (1:500; Pharmingen); anti-E2F1 rabbit polyclonal antibody C-20 (1:300; Santa Cruz); anti-mouse hemoglobin rabbit polyclonal antibody (1:1,000; ICN); and anti-cyclin A rabbit polyclonal antibody C19 (1:500; Santa Cruz). Horseradish peroxidase-conjugated secondary antibodies included anti-mouse antibody (1:5,000; Sigma) and anti-rabbit antibody (1:3,500; Santa Cruz). Horseradish peroxidase activity was detected by using the enhanced chemiluminescence (ECL) system (Amersham).
RESULTS
PU.1 represses GATA-1 transcriptional activity by binding to it on DNA.
As mentioned above, PU.1 has been reported to inhibit the in vitro binding of GATA-1 to DNA (39, 72), but the significance of such inhibition for PU.1-mediated repression of GATA-1 in vivo is unclear. Using electrophoretic mobility shift assays (EMSAs), we also found that PU.1 can inhibit GATA-1 DNA binding in vitro; however, by using a different in vitro assay, DNase I footprinting, we found that inhibition was strongly dependent on PU.1 concentration. Inhibition of the GATA-1 footprint was seen only at very high concentrations of PU.1 (>20-fold molar excess to GATA-1). At lower concentrations (approximately molar with GATA-1), PU.1 actually altered the GATA-1 generated footprint surrounding the GATA-1 binding site (N. Rekhtman, A. I. Skoultchi, Tianyuan Zhou, and Cheng-Ming Chiang, unpublished results). The latter observation is consistent with the possibility that PU.1 can bind to GATA-1 on DNA.
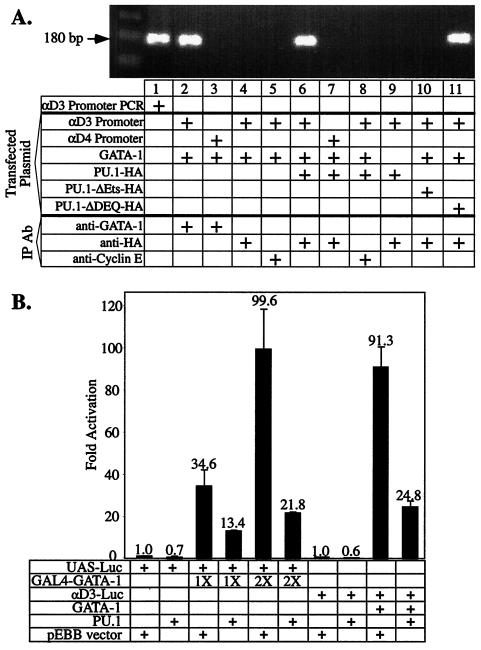
To determine whether PU.1 can bind to GATA-1 in vivo while it is on DNA, we performed ChIP experiments with a GATA-1-responsive luciferase reporter gene, αD3 Luc, driven by an α-globin promoter. We showed previously that transactivation of this reporter by GATA-1 is strongly repressed by PU.1 (50). We transfected the reporter, along with various combinations of expression vectors encoding GATA-1 and HA-tagged full-length PU.1 or mutants of PU.1, and then immunoprecipitated formaldehyde cross-linked chromatin with anti-GATA-1 or anti-HA antisera or an irrelevant antiserum (Fig. 1A). We found that the α-globin promoter sequences coimmunoprecipitated with both GATA-1 and PU.1 (Fig. 1A, lanes 2 and 6). Importantly, the promoter sequences did not coprecipitate with PU.1 unless GATA-1 is also expressed in the cells (lane 9). A second key control was the use of a reporter in which the GATA-1 binding sites in the promoter are mutated (αD4 Luc). In this case, the promoter sequences did not coimmunoprecipitate with GATA-1 or PU.1 when they were coexpressed (lanes 3 and 7). Very similar results were obtained with an anti-PU.1 antibody (data not shown). Consistent with our earlier findings (50), PU.1 deleted of its Ets domain, which is required for it to bind to GATA-1, cannot bind to GATA-1 on DNA (lane 10). However, PU.1 deleted of its N-terminal domain, which is not required for it to bind to GATA-1 but is required for it to repress GATA-1, can bind to GATA-1 on DNA (lane 11). As shown below, even though this mutant binds to GATA-1 on DNA, it fails to repress transcription because it cannot bind the corepressor pRB. We conclude that, in vivo, PU.1 can bind to GATA-1 when GATA-1 is on DNA. The results discussed below also show that PU.1 is present along with GATA-1 at an endogenous GATA-1 binding site in undifferentiated MEL cells.
FIG. 1.
PU.1 binds to GATA-1 on DNA. (A) A total of 107 U2OS cells were transfected by the Lipofectamine Plus method (Gibco-BRL) with 12.9 μg of DNA consisting of 420 ng of αD3-Luc, 2.8 μg of pXM-GATA-1, 2.8 μg of the indicated pEBB-PU.1-HA expression plasmid, and the required amount of pBSK vector DNA. ChIPs were carried out as described in Materials and Methods with anti-HA, anti-GATA-1, or anti-cyclin E antibodies. The presence of DNA sequences encompassing the GATA-1 site in the αD3 promoter (or the mutant GATA-1 site in the αD4 promoter) was determined by PCR assays as described in Materials and Methods. The presence of the 180-bp fragment indicates presence of the promoter region in the immunoprecipitate. Lane 1 shows PCR amplification of αD3-Luc plasmid DNA. A schematic diagram of the mutant PU.1 proteins used is shown in Fig. 3. (B) U2OS cells were transfected with a UAS-Luc reporter (100 ng) or the αD3-Luc reporter (15 ng), and pRSV-GATA-1-GAL4(DBD) (50 and 100 ng), pXM-GATA-1 (50 ng), and 30 ng of pEBB-PU.1 or 30 ng of empty pEBB vector as indicated. Luciferase activity was determined 48 h after transfection.
To further investigate whether PU.1 can repress GATA-1-activated transcription without disrupting DNA binding, we utilized a protein in which the C finger region of GATA-1 is replaced by sequences that direct GAL4 DNA binding to upstream activating sequences (UAS). Since PU.1 does not interact with the GAL4 DNA-binding region, PU.1 cannot interfere with binding of the chimera to UAS (data not shown). PU.1 repressed transcriptional activation by the chimeric protein as well as it repressed GATA-1 itself (Fig. 1B). These experiments further support the view that PU.1 can repress GATA-1-mediated transcriptional activity without interfering with its binding to DNA.
PU.1 and pRB interact in vitro and in vivo and colocalize with GATA-1 at GATA-1 binding sites.
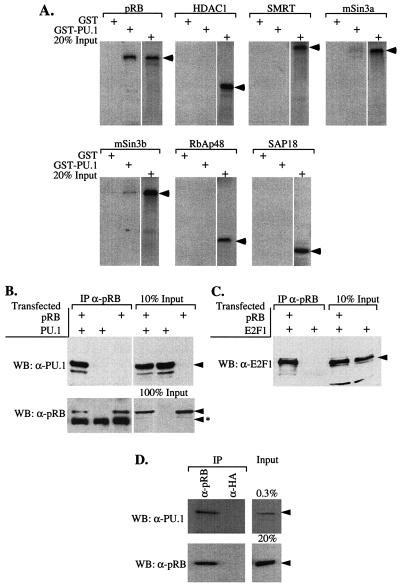
The finding that PU.1 can bind to GATA-1 on DNA even without its N-terminal region (Fig. 1A), but that such binding does not inhibit GATA-1 transcriptional activity (50), suggested that PU.1 might require a cofactor to cause repression. The N-terminal region of PU.1 was reported previously to bind pRB (16, 29). Therefore, we tested the in vitro binding of PU.1 to a set of previously identified transcriptional corepressors, including pRB. GST-PU.1 bound pRB with apparent high affinity (Fig. 2; see also Fig. 4). Weak interactions also were detected with Sin3a and Sin3b (Fig. 2A) and the pRB-related pocket protein, p107 (data not shown). No interaction was detected with HDAC1, SMRT, RbAp48, or SAP18.
FIG. 2.
PU.1 and pRB interact in vitro and in vivo. (A) 35S-labeled proteins, indicated above each panel, were prepared by coupled transcription-translation reactions and tested for interaction with GST or GST-PU.1 immobilized on glutathione-Sepharose beads as described in Materials and Methods. Bound proteins were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and autoradiography. An autoradiogram of 20% of each 35S-labeled protein added to the binding reactions is shown in each panel. Arrowheads indicate the positions of the 35S-labeled proteins on the gel. (B and C) Extracts of 293 cells, transfected with expression constructs for PU.1, E2F1, and pRB (large pocket, residues 379 to 928) as indicated, were immunoprecipitated (IP) with anti-pRB (XZ91; Pharmingen) antibody. Immunoprecipitated complexes were resolved by SDS-PAGE and analyzed by Western blotting (WB) with the indicated primary antibodies. Arrowheads mark the position of specifically immunodetected proteins; arrowheads with an asterisk indicate the position of the immunoglobulin chains used in the immunoprecipitaions. An immunoblot of the indicated fraction of each type of extract added to the immunoprecipitation reactions is shown in each panel (% input lanes). (D) Extracts of MEL cells were immunoprecipitated with anti-pRB (G3-245; Pharmingen) antibody and analyzed as in panel B.
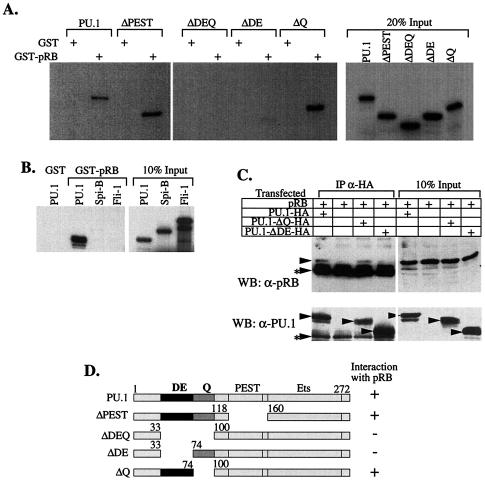
FIG. 4.
The acidic subdomain of PU.1 is required for interaction of PU.1 with pRB in vitro and in vivo. (A and B) 35S-labled proteins prepared by coupled transcription-translation reactions were tested for interaction with GST or GST-pRB as described in Fig. 2. (C) 293 cells were transfected with pRB (large pocket, residues 379 to 928) and expression vectors encoding PU.1 or PU.1-mutant proteins tagged with an HA epitope. Cell extracts were prepared and immunoprecipitated (IP) with anti-HA antibody. Bound complexes were analyzed by Western blotting with anti-pRB (XZ55) or anti-PU.1 primary antibodies. Other details are as in Fig. 2. (D) Schematic diagram of the wild-type and mutant PU.1 proteins used in the interaction studies in panels A to C and a summary of the results for PU.1 interaction with pRB. DE and Q indicate the acidic and glutamine residue-rich subdomains, respectively; PEST, region rich in PEST residues; Ets, DNA-binding Ets homology domain.
To test whether PU.1 and pRB associate in vivo, we performed coimmunoprecipitation experiments with extracts of 293 cells transfected with expression vectors encoding PU.1 and pRB. PU.1 was readily detected in the anti-pRB immunoprecipitates (Fig. 2B; see also Fig. 4 for anti-PU.1 immunoprecipitates). The apparent affinity of PU.1 and pRB was similar to that of pRB and two of its established binding partners, E2F1 (Fig. 2C) and E1A (data not shown). PU.1-pRB complexes also could be observed in U20S and C33A cells transfected with the expression constructs (data not shown). An association of PU.1 with endogenous pRB in 293 cells also was seen, after prolonged exposures of the anti-PU.1 immunoblots (data not shown).
To determine whether endogenous PU.1 and pRB associate in MEL cells, we analyzed pRB immunoprecipitates for PU.1 by immunoblotting. A complex of PU.1 and pRB was readily detected in MEL cells (Fig. 2D). pRB is present in both hypo- and hyperphosphorylated forms in MEL cells. When GST-PU.1 was exposed to a MEL cell extract, it preferentially bound hypophosphorylated pRB (data not shown).
ChIP experiments presented in the previous section show that PU.1 binds to GATA-1 on a transfected α-globin promoter-reporter gene. We used ChIP to determine whether pRB colocalizes with PU.1 and GATA-1 on the α-globin promoter. We transfected the reporter, along with various combinations of expression vectors encoding one of the three proteins and then performed ChIP with either anti-RB antiserum or a control antiserum. We found that pRB is present on the promoter only when PU.1 and GATA-1 are also expressed in the cells (Fig. 3A). Furthermore, binding to the mutated αD4 promoter did not occur (Fig. 3A, lane 4). These results indicate that a GATA-1-PU.1-pRB complex can form at a functional GATA-1 binding site in DNA and that formation of the complete complex is dependent on the presence of GATA-1 and PU.1 and an intact GATA-1 DNA-binding site.
Several studies have suggested that GATA-1 is present at GATA-1 binding sites in undifferentiated MEL cells (1, 10, 20, 48, 56, 68). Recently, ChIP experiments demonstrated that GATA-1 strongly localizes near the single GATA-1 binding site in the DNase I hypersensitive site-2 (HS2) in the β-globin locus control region in differentiated MEL cells (23). We used ChIP to determine whether PU.1 and pRB are present along with GATA-1 near HS2 in undifferentiated MEL cells. The amounts of the HS2 DNA fragment in immunoprecipitated chromatin were compared by quantitative PCR to that of the G6PD gene promoter that does not contain known GATA-1 binding sites and an enrichment factor was calculated. The Ey embryonic globin gene promoter that is inactive in MEL cells was used as a negative control. As shown in Fig. 3B, GATA-1 occupies the HS2 region in MEL cells. ChIP with anti-PU.1 and anti-pRB antibodies show that PU.1 and pRB also occupy the same region. None of the three proteins were found to occupy the Ey promoter region. These results show that PU.1 and pRB colocalize with GATA-1 at an endogenous GATA-1 target site in undifferentiated MEL cells in which PU.1 functions to inhibit GATA-1 and block erythroid differentiation.
Interaction of PU.1 and pRB is mediated by the acidic subdomain of PU.1 and the C pocket of pRB.
To define regions within PU.1 and pRB that are required for their interaction, we tested a set of deletion mutants of each protein for their abilities to interact in vitro. The defined functional domains of PU.1 include an N-terminal region that is needed for transcriptional transactivation. This region includes acidic and glutamine-rich subdomains (Fig. 4D), both of which participate in transactivation. There is also a central region rich in proline, glutamic acid, serine, and threonine (PEST) residues and a C-terminal Ets homology region that binds DNA and also GATA-1. We found that the acidic subdomain within the N-terminal region of PU.1 was specifically required for binding to GST-pRB in vitro (Fig. 4A). The requirement for this subdomain for binding to pRB was also observed in vivo (Fig. 4C).
PU.1 is a member of the Ets family of transcription factors that share homology in the Ets region responsible for DNA binding. However, these proteins diverge significantly in other regions. The PU.1 Ets region is most closely related to that of Spi-B, a B-cell-specific factor (47). Fli-1 is another member of the family that, like PU.1, is involved in certain virus-induced erythroleukemias (3). pRB interacted selectively with PU.1 (Fig. 4B), a finding consistent with the localization of binding to the PU.1 acidic subdomain and the divergence of the three proteins in their N-terminal regions.
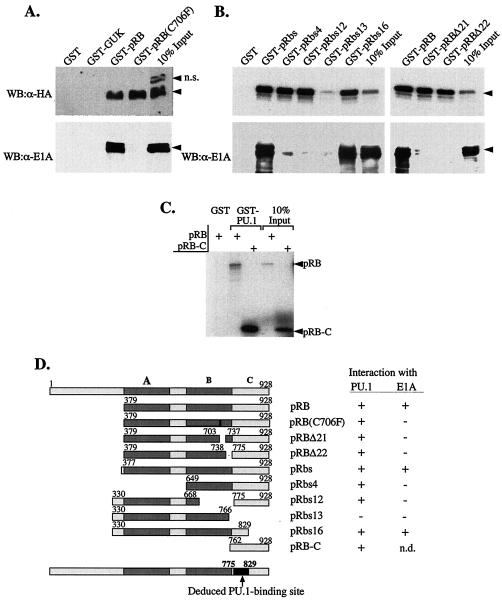
The domains of pRB include an important C-terminal region consisting of the so-called A, B, and C domains (30). Many pRB-interacting proteins bind within this region, and most of these interactions depend on the so-called A/B pocket. To identify the region within pRB mediating interaction with PU.1, we tested a panel of GST-pRB mutants for binding to PU.1. As a control we also tested binding of the same pRB mutants to E1A, a highly studied pRB interacting protein. Deletion or disruption of the structure of the A/B pocket, e.g., by mutation at pRB residue 706, causes a loss of E1A binding (24, 25). We found that such mutations did not affect PU.1 binding to pRB. On the other hand, deletion of the pRB C domain caused a nearly complete loss of PU.1 binding (Fig. 5A and B). Furthermore, the pRB C domain bound to GST-PU.1 as well as the full-length protein pRB (Fig. 5C). Thus, the C domain of pRB is both necessary and sufficient for interaction with PU.1. The results of studies with mutant pRbs16 suggest that the PU.1 binding region lies between residues 775 and 829 in pRB.
FIG. 5.
The C pocket of pRB mediates interaction with PU.1. (A) An expression construct encoding PU.1-HA was transfected into 293 cells, which endogenously express E1A, and cell extracts were tested for interaction with GST or the GST fusion proteins indicated above the panel. Bound proteins were analyzed by SDS-PAGE and Western blot analysis (WB) with anti-HA or anti-E1A primary antibodies. Arrowheads mark the position of the specifically immunodetected proteins. n.s., Position of a nonspecific band detected in 293 cell extracts with anti-HA antibody. (B and C) GST or the GST fusion proteins indicated above each panel were tested for interaction with 35S-labeled PU.1 made in coupled transcription-translation reactions, and the bound complexes were analyzed by SDS-PAGE and autoradiography. In panel B (lower), the same amounts of GST proteins were incubated with 293 cell extracts, and bound E1A was analyzed as in panel A. (D) Schematic diagram of the wild-type and mutant pRB proteins used and a summary of binding results. A-, B-, and C-pocket regions of pRB are indicated as A, B, and C, respectively. The deduced region of pRB that interacts with PU.1 is also shown.
The pRB-binding, acidic subdomain of PU.1 is required for repression of GATA-1 and inhibition of erythroid differentiation.
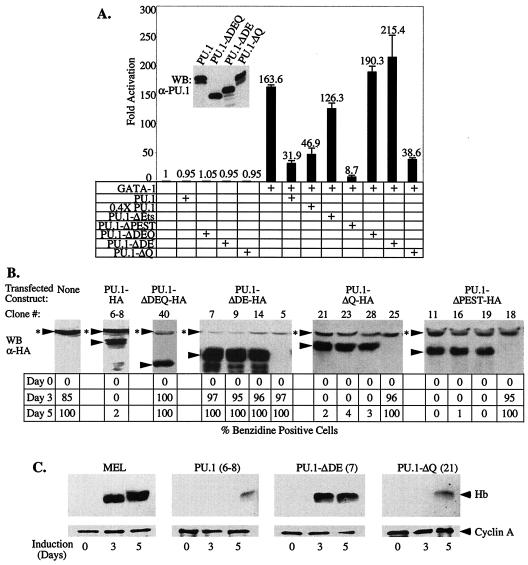
Having established that the acidic subdomain of PU.1 is required for it to bind pRB, a known transcriptional corepressor, we next sought to determine whether this region is required for PU.1 to repress GATA-1 transcriptional activity. The ability of various PU.1 deletion mutants to repress GATA-1-stimulated transcription of the αD3 Luc reporter was tested by cotransfection experiments (Fig. 6A). As reported previously, deletion of the Ets region reduced PU.1-mediated repression of GATA-1 because this region is required for binding to GATA-1. Deletion of the PEST region had no effect on repression. Deletion of either residues 34 to 99 or the acidic subdomain, residues 34 to 73 (Fig. 4D), eliminated repression (Fig. 6A). Importantly, deletion of the glutamine-rich subdomain adjacent to the acidic region had no effect on repression. Thus, the acidic subdomain of PU.1 is required both for binding pRB and for repression of GATA-1.
FIG. 6.
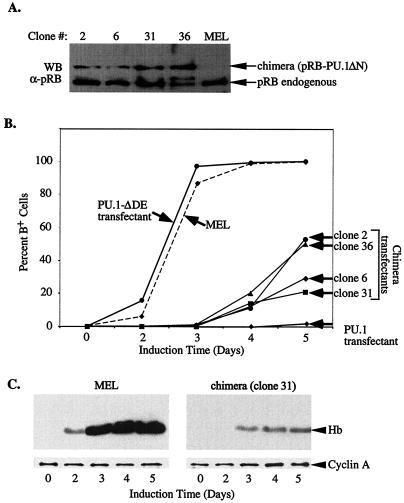
The acidic subdomain of PU.1 is required for repression of GATA-1 and inhibition of MEL cell differentiation. (A) U2OS cells were transfected with the αD3-Luc reporter (15 ng), pXM-GATA-1 (50 ng), and 100 ng of pEBB-PU.1 expression constructs or empty pEBB vector, as indicated. Luciferase activity was determined 48 h after transfection. (Inset) The expression level of the indicated proteins was analyzed by Western blotting (WB) with an anti-PU.1 antibody. (B) Stably transfected MEL cells clones expressing the indicated HA epitope-tagged PU.1 proteins were generated as described in Materials and Methods. The levels of transfected proteins were determined by Western blot analysis (WB) with an anti-HA antibody and compared to previously described tranfectants expressing wild-type PU.1-HA (clone 6-8) and PU.1-ΔDEQ-HA (50). Three clones with representative expression levels are shown for each type of expression construct. Arrowheads mark the position of specifically immunodetected proteins; arrowheads with asterisks indicate the position of a nonspecific band detected with the anti-HA antibody. The clones were treated with 1.8% DMSO and, at the indicated times, the percentage of hemoglobinized, benzidine-positive cells was determined. Similar results were obtained with three to five additional clones for each type of transfected construct. (C) Cell extracts of the indicated transfectants (clone number in parenthesis) were prepared after 3 and 5 days of treatment with 1.8% DMSO and from untreated cells (day 0). Equal amounts of protein were loaded in each lane and hemoglobin (Hb) levels were determined by Western blot analysis with an anti-Hb antibody. An anti-cyclin A antibody was used as a loading control. A schematic diagram of the mutant PU.1 proteins is shown in Fig. 4D.
PU.1 is also able to block erythroid differentiation. We have presented evidence that this activity is very likely due at least in part to its ability to repress GATA-1 (50). We have described previously an assay for the activity of PU.1 in blocking erythroid differentiation based on its ability to inhibit chemically induced differentiation of MEL cells (46). Treatment of MEL cells with agents such as dimethyl sulfoxide (DMSO) leads to a decline in endogenous PU.1 levels, permitting the cells to reenter their differentiation program, culminating in the accumulation of hemoglobin. In MEL cells that have been stably transfected with an expression vector encoding PU.1, however, such treatment does not lead to reduction in exogenous PU.1 levels, and hence the cells are inhibited from differentiating. We tested the ability of various deletion mutants of PU.1 to inhibit MEL cell differentiation by generating MEL cell lines expressing HA epitope-tagged wild-type and mutant PU.1 proteins. Transfectants expressing amounts of the mutant proteins at levels similar to or greater than previously characterized wild-type PU.1 transfectants were identified by immunoblotting with an anti-HA antibody (Fig. 6B). The transfectants were then tested for their ability to undergo DMSO-induced differentiation as measured by the production of hemoglobinized cells by staining with benzidine reagent (Fig. 6B) and by production of hemoglobin by Western blotting (Fig. 6C). As reported previously, transfectants expressing exogenous, wild-type PU.1 at appropriate levels (approximately equal to endogenous PU.1) (46, 50), are nearly completely blocked from differentiating, whereas transfectants expressing a PU.1 mutant deleted of residues 34 to 99 differentiate well (Fig. 6B and C). Deletion of the glutamine-rich subdomain had little or no effect on the ability of PU.1 to block differentiation. However, the PU.1 mutant deleted of the acidic subdomain was unable to inhibit differentiation, despite being expressed at consistently higher levels than wild-type PU.1 or other mutant PU.1 proteins (Fig. 6B and C). Thus, the PU.1 acidic subdomain required for pRB binding and repression of GATA-1 is also required for blocking erythroid differentiation.
pRB is required for PU.1-mediated repression of GATA-1 and inhibition of erythroid differentiation.
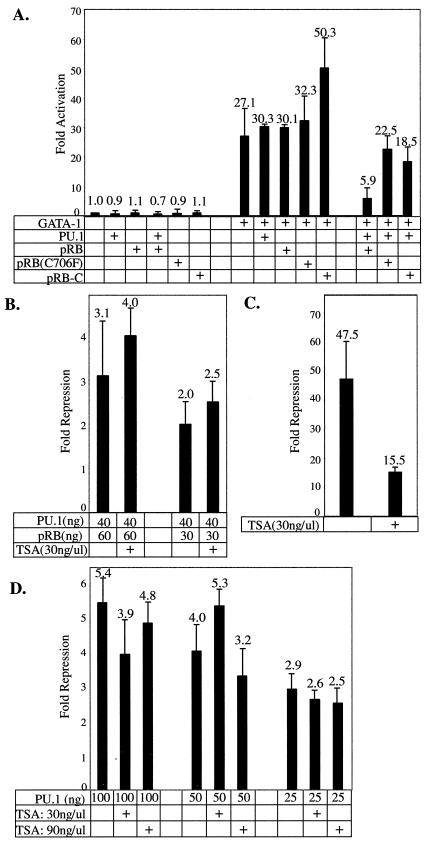
The foregoing results indicate that the PU.1 acidic subdomain is required for pRB binding, as well as for PU.1-mediated repression of GATA-1 and inhibition of erythroid differentiation. To test whether pRB is actually required for repression of GATA-1 by PU.1, we performed reporter assays in pRB-null SAOS2 osteosarcoma cells. We found that PU.1 was unable to repress GATA-1-stimulated transcription in SAOS2 cells (Fig. 7A), whereas it can repress GATA-1 in the related, pRB-positive osteosarcoma cells, U2OS (Fig. 6A). Expression of pRB along with PU.1 in SAOS2 cells restored its ability to repress GATA-1. Importantly, pRB did not affect GATA-1 stimulated transcription in the absence of PU.1 (Fig. 7A). PU.1 also was unable to repress GATA-1 in pRB-null C33A cells and repression was restored by expression of pRB (data not shown). Repression also was not observed in 293 cells, in which pRB is inactivated by the adenovirus E1A protein. We conclude that PU.1-mediated repression of GATA-1 requires participation of pRB.
FIG. 7.
pRB is required for PU.1-mediated repression of GATA-1. SAOS2 cells were transfected with 25 ng of αD3-Luc reporter, 50 ng of pXM-GATA-1, 40 ng of pEBB-PU.1, and 60 ng of cytomegalovirus promoter-driven expression constructs encoding pRB or pRB mutants as indicated. Equivalent amounts of pEBB and pCMV expression constructs were used in each transfection by including empty vectors as needed. (B) SAOS2 cells were transfected as in panel A. TSA was added 24 h after transfection was initiated, and the luciferase activity was assayed 24 h later. (C) SAOS2 cells were transfected with 100 ng of UAS-Luc reporter, 50 ng of GAL4-E2F1, and 60 ng of pCMV-pRB expression vector with or without TSA addition, as indicated. (D) U2OS cells were transfected as in Fig. 6A, with or without TSA addition, as indicated.
Since the pRB C domain is both necessary and sufficient for interaction with PU.1 (Fig. 5), we sought to determine whether it is sufficient for PU.1-mediated repression of GATA-1. We observed very little repression with the C domain fragment (Fig. 7A), suggesting that other regions of pRB are needed for repression. Consistent with this view, we also observed very little repression with the C706F mutant of pRB (Fig. 7A), which disrupts the structure of the A/B pocket but does not affect pRB binding to PU.1.
Histone deacetylases (HDACs) have been implicated in some instances of pRB-mediated repression (6, 35, 37), but HDAC-independent mechanisms also have been documented (35, 40, 52). We found that TSA, a specific chemical inhibitor of some HDACs, did not reverse PU.1-mediated repression of GATA-1 in SAOS2 cells (Fig. 7B), U2OS cells (Fig. 6D), and HeLa cells (data not shown). TSA did reverse pRB-mediated repression of E2F1 in SAOS2 cells (Fig. 7C), as previously reported (37). Therefore, repression of GATA-1-dependent transactivation of the α-globin promoter by PU.1 and pRB appears to be independent of HDAC activity in three cell lines.
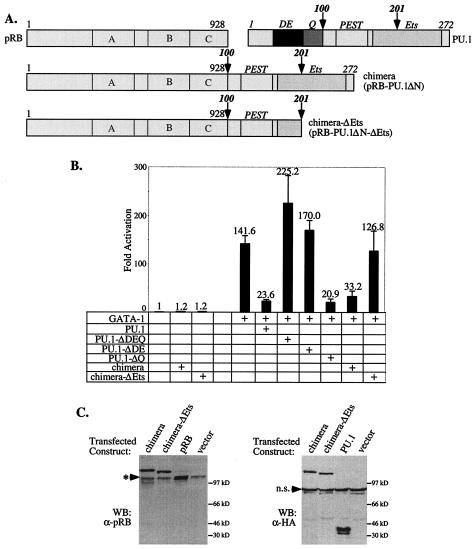
As discussed above, in addition to repressing GATA-1 transcriptional activity, PU.1 also can inhibit differentiation of MEL cells. To test whether this function of PU.1 is also dependent on pRB, we took advantage of our finding that deletion of the PU.1 N-terminal region, to which pRB binds, destroys PU.1 activity in both repression and inhibition of differentiation. Therefore, we sought to determine whether fusing pRB to the deleted PU.1 protein would restore its activity. We generated an expression vector (pRB-PU.1ΔN) in which the complete pRB coding sequence was placed upstream of PU.1 coding sequences specifying residues 100 to 272, which were followed by sequences encoding the HA epitope tag (Fig. 8A). As a control, we also constructed a vector (pRB-PU.1ΔN-ΔEts) encoding a chimeric pRB-PU.1 protein also lacking a portion of the PU.1 Ets region that is required for binding PU.1 to GATA-1 (Fig. 8A). Fusion proteins of the expected sizes were produced upon transfection of these expression constructs (Fig. 8C).
FIG. 8.
Fusion of pRB to a mutant PU.1 lacking the N-terminal region restores its ability to repress GATA-1. (A) Schematic diagram of the pRB:PU.1 fusion proteins used. Full-length pRB was fused to the amino terminus of PU.1 amino acid residues 100 to 272 (chimera; pRB-PU.1ΔN) or 100 to 201 (chimera-ΔEts; pRB-PU.1ΔN-ΔEts). Both chimeric constructs encode a C-terminal HA tag. (B) U2OS cells were transfected with the αD3-Luc reporter and the indicated expression constructs as in Fig. 6A and analyzed for luciferase activity. (C) Expression of the fusion proteins was verified by Western blotting (WB) with the indicated primary antibodies. Migration of the endogenous pRB is marked by an arrowhead with an asterisk. n.s., nonspecific band detected with anti-HA antibody.
We first tested the activity of the pRB-PU.1 fusion proteins for their ability to repress GATA-1. Fusion of pRB to PU.1 deleted of its N-terminal region restored its ability to inhibit GATA-1 (Fig. 8B). As expected, restoration of activity by fusion with pRB requires an intact Ets region because this region is needed for binding the fusion protein to GATA-1. These observations provide additional support for the view that PU.1 represses GATA-1 by binding to and tethering pRB to GATA-1.
To determine whether fusion of pRB to the N-terminal deleted PU.1 restored its ability to inhibit differentiation, we generated stable MEL cell lines expressing the pRB-PU.1ΔN fusion protein. We screened for expression by immunoblotting and selected several clones with the highest levels of the fusion protein. Immunoblotting showed that the levels of the fusion protein in these clones were lower than endogenous pRB (Fig. 9A) and also lower than the levels of exogenous full-length PU.1-HA produced in most transfectants that are blocked from differentiating (e.g., clone 6-8 in Fig. 6B) (data not shown). Despite the reduced expression levels of the fusion protein in these clones, they were inhibited from differentiating, compared to MEL cells or transfectants expressing PU.1 deleted of its pRB binding domain (Fig. 9B and C). However, as judged by benzidine staining, a sensitive indicator of hemoglobin levels in cells, the block to erythroid differentiation imposed by the pRB-PU.1 fusion protein is not as strong as that achieved with full-length PU.1. After 5 days of DMSO treatment some of the clones produced significant numbers of benzidine-positive cells, although hemoglobin levels in the cells were still quite low as measured by immunoblotting (Fig. 9C). The difference between the two types of clones may be attributable to the lower expression levels of the fusion protein in MEL cell transfectants or to the possible involvement of other corepressors or other functions of PU.1 that are not carried out by the pRB-PU.1ΔN fusion protein (see Discussion).
FIG. 9.
Fusion of pRB to mutant PU.1 lacking the N-terminal region restores its ability to inhibit MEL cell differentiation. (A) MEL cells were transfected with expression constructs encoding both resistance to puromycin and the pRB-PU.1 fusion protein (pRB-PU.1ΔN). Puromycin-resistant clones were isolated and analyzed for expression of the fusion protein by Western blotting (WB) of cell extracts with anti-pRB antibody (G3-245). (B) MEL cell clones expressing the pRB-PU.1 fusion protein shown in panel A were treated with 1.8% DMSO, and the percentage of benzidine-positive cells was assessed at the indicated times. Control cell lines used were MEL cells, a transfectant expressing a mutant PU.1 lacking a portion of the N-terminal region (PU.1-ΔDE, clone 7, Fig. 6B) and a transfectant expressing full-length PU.1 (PU.1 transfectant, clone 6-8, Fig. 6B). (C) Hemoglobin (Hb) production was determined as in Fig. 6C.
DISCUSSION
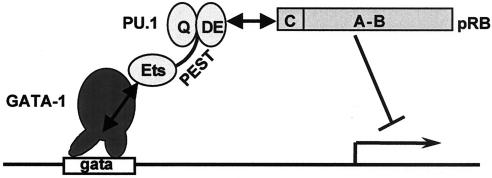
Previous work from several laboratories has indicated that a mutual antagonism between PU.1 and GATA-1 can affect lineage-specific differentiation decisions during hematopoietic development (39, 43, 50, 72). Moreover, the ability of PU.1 to inhibit GATA-1 is central to the block to differentiation in cultured erythroleukemic cells and is likely to contribute strongly to the development of erythroleukemia in Friend leukemia virus-infected mice. The results reported here suggest a mechanism by which PU.1 antagonizes the action of GATA-1. By binding directly to GATA-1 on DNA, PU.1 tethers to it an established transcriptional corepressor, pRB, thereby blocking GATA-1-mediated transcriptional activation and its ability to promote erythroid differentiation (Fig. 10). Evidence supporting this model consists of the following observations. (i) ChIP experiments indicate that PU.1 and pRB can be found associated with a GATA-1 target gene, but only when both GATA-1 and an intact GATA-1 DNA binding site are present. The three proteins also were found to colocalize on an endogenous GATA-1 binding site in MEL cells that are blocked from differentiating due to expression of PU.1. (ii) PU.1 and pRB interact in vitro and in vivo. A small acidic N-terminal region in PU.1 (the acidic DE subdomain) directs interaction with the C pocket of pRB. (iii) The acidic subdomain of PU.1 is required for its ability to repress GATA-1 and inhibit erythroid differentiation. (iv) PU.1 cannot repress GATA-1-mediated transcription in pRB-null cells, and pRB can restore repression. Fusion of pRB to a defective PU.1 mutant also restores its ability to repress GATA-1 and to inhibit erythroid differentiation.
FIG. 10.
Model of PU.1-mediated repression of GATA-1. Protein-protein interactions are indicated by double-headed arrows. The figure indicates that the Ets domain of PU.1 binds to the GATA-1 zinc finger region while it is bound to DNA. The acidic subdomain (DE) of PU.1 interacts with the C pocket of pRB. Tethering of pRB by PU.1 leads to repression of transcription.
The model proposed here for PU.1 inhibition of GATA-1 function does not exclude contributions from other mechanisms that have been proposed. For example, PU.1 has been proposed to inhibit GATA-1 function by blocking GATA-1 binding to DNA (39, 72). The evidence in favor of this mechanism is derived exclusively from studies by using in vitro EMSAs. It is possible that at very high concentrations of PU.1, GATA-1 binding to DNA is inhibited. However, we found that both PU.1 and GATA-1 can be readily detected on a GATA-1 target gene in cells in which GATA-1 transcriptional activity is repressed by PU.1. Moreover, we also found that PU.1 can repress activity of a GAL4-GATA-1 fusion protein that does not depend on the GATA-1 DNA-binding domain for transcriptional activity. In addition, several studies have suggested that GATA-1 is present at erythroid-specific promoters in undifferentiated mouse erythroleukemia cells (1, 10, 20, 48, 56, 68). Since these cells underwent malignant transformation and acquired a block to differentiation due to deregulated expression of PU.1, very likely they have much higher levels of PU.1 than normal erythroid-myeloid precursors in which PU.1 and GATA-1 may be coexpressed. Thus, the physiological relevance of PU.1 inhibition of GATA-1 DNA binding remains to be determined.
PU.1 also has been demonstrated to inhibit protein acetylation carried out by the acetyltransferase (AT) coactivator CBP. CBP can serve as a coactivator with GATA-1, and PU.1 inhibits CBP-mediated acetylation of GATA-1 (20). It has been suggested that such inhibition might contribute to the PU.1-mediated block to differentiation in MEL cells. However, a PU.1 deletion mutant lacking the N-terminal region (residues 33 to 100), including the acidic subdomain, retained substantial activity for inhibiting CBP-mediated protein acetylation. Because this mutant is completely inactive in repressing GATA-1 and in blocking erythroid differentiation, further studies are needed to assess the role of PU.1-mediated inhibition of CBP AT activity in these processes. Nevertheless, partial loss of inhibitory activity against the CBP AT might help to explain why we found that the pRB-PU.1ΔN fusion protein has reduced activity in blocking MEL cell differentiation compared to intact, full-length PU.1.
We found that PU.1 also can interact in vitro with several other corepressors, including Sin3a and Sin3b and p107. These interactions appeared to be much weaker than that of PU.1 and pRB. PU.1 was reported previously to interact directly with Sin3a and thereby indirectly with HDAC1 and to cause repression of several promoters (26). However, a PU.1 deletion mutant lacking residues 1 to 100 was also found to interact with Sin3a. Since this mutant is inactive in repressing GATA-1 and inhibiting erythroid differentiation, it seems unlikely that Sin3a plays an important role in these processes. In addition, since PU.1 was unable to repress GATA-1 in SAOS2 and C33A cells that contain p107, and since pRB was able to fully restore repression, it is less likely that p107 plays a role in the PU.1-mediated block to erythroid differentiation. In contrast to PU.1 levels that decline within 1 to 2 days after chemical induction of MEL differentiation is initiated, the level of p107 actually rises significantly during this period (data not shown), further suggesting that p107 may not have a role in the PU.1-mediated block to differentiation in these cells.
Extensive studies indicate that pRB plays a major role in the control of cell proliferation. pRB is thought to inhibit the cell cycle by negatively regulating E2F transcription factors that stimulate the expression of genes required for cell cycle progression (18). pRB has been shown to mediate transcriptional repression by recruiting to promoters several different types of chromatin remodeling activities, including enzymes that catalyze posttranslational modifications of histones, specifically histone deacetylases and histone methyltransferase, as well as complexes that alter the structure of nucleosomes (6, 35, 37, 51, 69, 70). We found that PU.1-mediated repression of GATA-1 was not reversed by TSA, suggesting that HDACs may not be involved. Several other instances of HDAC-independent pRB-mediated repression have been described (35, 40, 52). Of particular interest in the current context is the finding that pRB can interact with the corepressor CtBP and its interacting protein CtIP, which together can repress transcription in an HDAC-independent manner (40). CtBP has been reported to interact with a large number of vertebrate transcription factors (8), among them FOG-1 and FOG-2 that also interact with GATA factors. Thus, CtBP is an interesting candidate for involvement in HDAC-independent, PU.1-pRB-mediated repression of GATA-1. Interaction of FOG-1 and FOG-2 with CtBP has been reported to lead to repression of GATA-1 transcriptional activity (14, 19).
We have demonstrated that pRB interacts with a region of PU.1 required for repression of GATA-1 and for blocking erythroid differentiation and that pRB is involved in these processes. The involvement of pRB, which is both a key cell cycle inhibitor and transcriptional corepressor, suggests a dual mechanism for the PU.1-mediated inhibition of erythroid differentiation. On one hand, recruitment of pRB by PU.1 to GATA-1 target genes results in their repression. In addition, by sequestering a cell cycle inhibitor, PU.1 may also promote cell proliferation and tumorigenicity. Furthermore, pRB has also been shown to have differentiation-promoting functions (33), although in most instances its mechanism of action is not known. In at least one instance, however, pRB appears to function as a transcriptional coactivator by interacting with the CBFA1 transcription factor to promote osteoblast differentiation (59). pRB also is required for erythropoiesis during embryonic development. However, experiments with Rb−/−:Rb+/+ chimeric mice indicate that the principal function of pRB in erythropoiesis is not cell autonomous (36, 64). Nevertheless, certain aspects of erythropoiesis in such mice appear abnormal (21), and these observations have led to the suggestion that pRB may have a cell autonomous role in maturation of developing erythroblasts (63). If this is the case, then interaction of PU.1 with pRB might also affect its role in promoting the late stages of erythroid differentiation. The system described here may allow further insights into this important topic.
Acknowledgments
We are extremely grateful to Tianyuan Zhou and Cheng-Ming Chiang for carrying out the EMSA and footprinting experiments with PU.1 and GATA-1. We also thank Todd Evans, Richard Pestell, Leila Alland, and Liang Zhu for providing critical reagents and advice.
A.I.S. received support from National Cancer Institute Cancer Center grant 2P30CA13330. N.R., I.M., and K.C. were supported by NIH/MSTP 5T32GM07288-25. This work was supported by NIH grant 5R37CA16368.
I.M., S.M., and T.S. contributed equally to this study.
REFERENCES
- 1.Aumont, F. L., P. Trudel, and L. Wall. 1993. Murine erythroleukemia cells contain two distinct GATA-binding proteins that have different patterns of expression during cellular differentiation. Differentiation 52:169-176. [DOI] [PubMed] [Google Scholar]
- 2.Behre, G., A. J. Whitmarsh, M. P. Coghlan, T. Hoang, C. L. Carpenter, D. E. Zhang, R. J. Davis, and D. G. Tenen. 1999. c-Jun is a JNK-independent coactivator of the PU.1 transcription factor. J. Biol. Chem. 274:4939-4946. [DOI] [PubMed] [Google Scholar]
- 3.Ben-David, Y., and A. Bernstein. 1991. Friend virus-induced erythroleukemia and the multistage nature of cancer. Cell 66:831-834. [DOI] [PubMed] [Google Scholar]
- 4.Blobel, G. A., T. Nakajima, R. Eckner, M. Montminy, and S. H. Orkin. 1998. CREB-binding protein cooperates with transcription factor GATA-1 and is required for erythroid differentiation. Proc. Natl. Acad. Sci. USA 95:2061-2066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boyd, K. E., and P. J. Farnham. 1997. Myc versus USF: discrimination at the cad gene is determined by core promoter elements. Mol. Cell. Biol. 17:2529-2537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brehm, A., E. A. Miska, D. J. McCance, J. L. Reid, A. J. Bannister, and T. Kouzarides. 1998. Retinoblastoma protein recruits histone deacetylase to repress transcription. Nature 391:597-601. [DOI] [PubMed] [Google Scholar]
- 7.Cantor, A. B., and S. H. Orkin. 2002. Transcriptional regulation of erythropoiesis: an affair involving multiple partners. Oncogene 21:3368-3376. [DOI] [PubMed] [Google Scholar]
- 8.Chinnadurai, G. 2002. CtBP, an unconventional transcriptional corepressor in development and oncogenesis. Mol. Cell 9:213-224. [DOI] [PubMed] [Google Scholar]
- 9.Courey, A. J., and S. Jia. 2001. Transcriptional repression: the long and the short of it. Genes Dev. 15:2786-2796. [DOI] [PubMed] [Google Scholar]
- 10.Crossley, M., M. Merika, and S. H. Orkin. 1995. Self-association of the erythroid transcription factor GATA-1 mediated by its zinc finger domains. Mol. Cell. Biol. 15:2448-2456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Delgado, M. D., P. Gutierrez, C. Richard, M. A. Cuadrado, F. Moreau-Gachelin, and J. Leon. 1998. Spi-1/PU.1 proto-oncogene induces opposite effects on monocytic and erythroid differentiation of K562 cells. Biochem. Biophys. Res. Commun. 252:383-391. [DOI] [PubMed] [Google Scholar]
- 12.Eisenbeis, C. F., H. Singh, and U. Storb. 1995. Pip, a novel IRF family member, is a lymphoid-specific, PU.1-dependent transcriptional activator. Genes Dev. 9:1377-1387. [DOI] [PubMed] [Google Scholar]
- 13.Fisher, R. C., and E. W. Scott. 1998. Role of PU.1 in hematopoiesis. Stem Cells 16:25-37. [DOI] [PubMed] [Google Scholar]
- 14.Fox, A. H., C. Liew, M. Holmes, K. Kowalski, J. Mackay, and M. Crossley. 1999. Transcriptional cofactors of the FOG family interact with GATA proteins by means of multiple zinc fingers. EMBO J. 18:2812-2822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guerrasio, A., G. Saglio, C. Rosso, A. Alfarano, C. Camaschella, F. Lo Coco, A. Biondi, A. Rambaldi, S. Nicolis, and S. Ottolenghi. 1994. Expression of GATA-1 mRNA in human myeloid leukemic cells. Leukemia 8:1034-1038. [PubMed] [Google Scholar]
- 16.Hagemeier, C., A. J. Bannister, A. Cook, and T. Kouzarides. 1993. The activation domain of transcription factor PU.1 binds the retinoblastoma (RB) protein and the transcription factor TFIID in vitro: RB shows sequence similarity to TFIID and TFIIB. Proc. Natl. Acad. Sci. USA 90:1580-1584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hannon, R., T. Evans, G. Felsenfeld, and H. Gould. 1991. Structure and promoter activity of the gene for the erythroid transcription factor GATA-1. Proc. Natl. Acad. Sci. USA 88:3004-3008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Harbour, J. W., and D. C. Dean. 2000. Rb function in cell-cycle regulation and apoptosis. Nat. Cell Biol. 2:E65-E67. [DOI] [PubMed] [Google Scholar]
- 19.Holmes, M., J. Turner, A. Fox, O. Chisholm, M. Crossley, and B. Chong. 1999. hFOG-2, a novel zinc finger protein, binds the co-repressor mCtBP2 and modulates GATA-mediated activation. J. Biol. Chem. 274:23491-23498. [DOI] [PubMed] [Google Scholar]
- 20.Hong, W., A. Y. Kim, S. Ky, C. Rakowski, S. B. Seo, D. Chakravarti, M. Atchison, and G. A. Blobel. 2002. Inhibition of CBP-mediated protein acetylation by the Ets family oncoprotein PU.1. Mol. Cell. Biol. 22:3729-3743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hu, N., M. L. Gulley, J. T. Kung, and E. Y. Lee. 1997. Retinoblastoma gene deficiency has mitogenic but not tumorigenic effects on erythropoiesis. Cancer Res. 57:4123-4129. [PubMed] [Google Scholar]
- 22.Huang, S., E. Shin, K. A. Sheppard, L. Chokroverty, B. Shan, Y. W. Qian, E. Y. Lee, and A. S. Yee. 1992. The retinoblastoma protein region required for interaction with the E2F transcription factor includes the T/E1A binding and carboxy-terminal sequences. DNA Cell Biol. 11:539-548. [DOI] [PubMed] [Google Scholar]
- 23.Johnson, K. D., J. A. Grass, M. E. Boyer, C. M. Kiekhaefer, G. A. Blobel, M. J. Weiss, and E. H. Bresnick. 2002. Cooperative activities of hematopoietic regulators recruit RNA polymerase II to a tissue-specific chromatin domain. Proc. Natl. Acad. Sci. USA 99:11760-11765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kaelin, W. G., Jr., M. E. Ewen, and D. M. Livingston. 1990. Definition of the minimal simian virus 40 large T antigen- and adenovirus E1A-binding domain in the retinoblastoma gene product. Mol. Cell. Biol. 10:3761-3769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kaelin, W. G., Jr., D. C. Pallas, J. A. DeCaprio, F. J. Kaye, and D. M. Livingston. 1991. Identification of cellular proteins that can interact specifically with the T/E1A-binding region of the retinoblastoma gene product. Cell 64:521-532. [DOI] [PubMed] [Google Scholar]
- 26.Kihara-Negishi, F., H. Yamamoto, M. Suzuki, T. Yamada, T. Sakurai, T. Tamura, and T. Oikawa. 2001. In vivo complex formation of PU.1 with HDAC1 associated with PU.1-mediated transcriptional repression. Oncogene 20:6039-6047. [DOI] [PubMed] [Google Scholar]
- 27.Klemsz, M. J., and R. A. Maki. 1996. Activation of transcription by PU.1 requires both acidic and glutamine domains. Mol. Cell. Biol. 16:390-397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Klemsz, M. J., S. R. McKercher, A. Celada, C. Van Beveren, and R. A. Maki. 1990. The macrophage and B cell-specific transcription factor PU.1 is related to the ets oncogene. Cell 61:113-124. [DOI] [PubMed] [Google Scholar]
- 29.Konishi, Y., M. Tominaga, Y. Watanabe, F. Imamura, A. Goldfarb, R. Maki, M. Blum, E. M. De Robertis, and A. Tominaga. 1999. GOOSECOID inhibits erythrocyte differentiation by competing with Rb for PU.1 binding in murine cells. Oncogene 18:6795-6805. [DOI] [PubMed] [Google Scholar]
- 30.Kouzarides, T. 1995. Transcriptional control by the retinoblastoma protein. Semin. Cancer Biol. 6:91-98. [DOI] [PubMed] [Google Scholar]
- 31.Kulessa, H., J. Frampton, and T. Graf. 1995. GATA-1 reprograms avian myelomonocytic cell lines into eosinophils, thromboblasts, and erythroblasts. Genes Dev. 9:1250-1262. [DOI] [PubMed] [Google Scholar]
- 32.Lin, R. J., T. Sternsdorf, M. Tini, and R. M. Evans. 2001. Transcriptional regulation in acute promyelocytic leukemia. Oncogene 20:7204-7215. [DOI] [PubMed] [Google Scholar]
- 33.Lipinski, M. M., and T. Jacks. 1999. The retinoblastoma gene family in differentiation and development. Oncogene 18:7873-7882. [DOI] [PubMed] [Google Scholar]
- 34.Look, A. T. 1997. Oncogenic transcription factors in the human acute leukemias. Science 278:1059-1064. [DOI] [PubMed] [Google Scholar]
- 35.Luo, R. X., A. A. Postigo, and D. C. Dean. 1998. Rb interacts with histone deacetylase to repress transcription. Cell 92:463-473. [DOI] [PubMed] [Google Scholar]
- 36.Maandag, E. C., M. van der Valk, M. Vlaar, C. Feltkamp, J. O'Brien, M. van Roon, N. van der Lugt, A. Berns, and H. te Riele. 1994. Developmental rescue of an embryonic-lethal mutation in the retinoblastoma gene in chimeric mice. EMBO J. 13:4260-4268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Magnaghi-Jaulin, L., R. Groisman, I. Naguibneva, P. Robin, S. Lorain, J. P. Le Villain, F. Troalen, D. Trouche, and A. Harel-Bellan. 1998. Retinoblastoma protein represses transcription by recruiting a histone deacetylase. Nature 391:601-605. [DOI] [PubMed] [Google Scholar]
- 38.Martin, D. I., and S. H. Orkin. 1990. Transcriptional activation and DNA binding by the erythroid factor GF-1/NF-E1/Eryf 1. Genes Dev. 4:1886-1898. [DOI] [PubMed] [Google Scholar]
- 39.Matsumura, I., A. Kawasaki, H. Tanaka, J. Sonoyama, S. Ezoe, N. Minegishi, K. Nakajima, M. Yamamoto, and Y. Kanakura. 2000. Biologic significance of GATA-1 activities in Ras-mediated megakaryocytic differentiation of hematopoietic cell lines. Blood 96:2440-2450. [PubMed] [Google Scholar]
- 40.Meloni, A. R., E. J. Smith, and J. R. Nevins. 1999. A mechanism for Rb/p130-mediated transcription repression involving recruitment of the CtBP corepressor. Proc. Natl. Acad. Sci. USA 96:9574-9579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Moreau-Gachelin, F., A. Tavitian, and P. Tambourin. 1988. Spi-1 is a putative oncogene in virally induced murine erythroleukaemias. Nature 331:277-280. [DOI] [PubMed] [Google Scholar]
- 42.Moreau-Gachelin, F., F. Wendling, T. Molina, N. Denis, M. Titeux, G. Grimber, P. Briand, W. Vainchenker, and A. Tavitian. 1996. Spi-1/PU.1 transgenic mice develop multistep erythroleukemias. Mol. Cell. Biol. 16:2453-2463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nerlov, C., E. Querfurth, H. Kulessa, and T. Graf. 2000. GATA-1 interacts with the myeloid PU.1 transcription factor and represses PU.1-dependent transcription. Blood 95:2543-2551. [PubMed] [Google Scholar]
- 44.Orkin, S. H. 2000. Diversification of haematopoietic stem cells to specific lineages. Nat. Rev. Genet. 1:57-64. [DOI] [PubMed] [Google Scholar]
- 45.Quang, C. T., M. Pironin, M. von Lindern, H. Beug, and J. Ghysdael. 1995. Spi-1 and mutant p53 regulate different aspects of the proliferation and differentiation control of primary erythroid progenitors. Oncogene 11:1229-1239. [PubMed] [Google Scholar]
- 46.Rao, G., N. Rekhtman, G. Cheng, T. Krasikov, and A. I. Skoultchi. 1997. Deregulated expression of the PU.1 transcription factor blocks murine erythroleukemia cell terminal differentiation. Oncogene 14:123-131. [DOI] [PubMed] [Google Scholar]
- 47.Ray, D., R. Bosselut, J. Ghysdael, M. G. Mattei, A. Tavitian, and F. Moreau-Gachelin. 1992. Characterization of Spi-B, a transcription factor related to the putative oncoprotein Spi-1/PU.1. Mol. Cell. Biol. 12:4297-4304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Reddy, P. M., and C. K. Shen. 1993. Erythroid differentiation of mouse erythroleukemia cells results in reorganization of protein-DNA complexes in the mouse beta maj globin promoter but not its distal enhancer. Mol. Cell. Biol. 13:1093-1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rekhtman, N. 2000. Functional antagonism of transcription factors PU.1 and GATA-1 in erythroleukemia cells. Albert Einstein College of Medicine, Bronx, N.Y.
- 50.Rekhtman, N., F. Radparvar, T. Evans, and A. I. Skoultchi. 1999. Direct interaction of hematopoietic transcription factors PU.1 and GATA-1: functional antagonism in erythroid cells. Genes Dev. 13:1398-1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Robertson, K. D., S. Ait-Si-Ali, T. Yokochi, P. A. Wade, P. L. Jones, and A. P. Wolffe. 2000. DNMT1 forms a complex with Rb, E2F1 and HDAC1 and represses transcription from E2F-responsive promoters. Nat. Genet. 25:338-342. [DOI] [PubMed] [Google Scholar]
- 52.Ross, J. F., X. Liu, and B. D. Dynlacht. 1999. Mechanism of transcriptional repression of E2F by the retinoblastoma tumor suppressor protein. Mol. Cell 3:195-205. [DOI] [PubMed] [Google Scholar]
- 53.Schuetze, S., R. Paul, B. C. Gliniak, and D. Kabat. 1992. Role of the PU.1 transcription factor in controlling differentiation of Friend erythroleukemia cells. Mol. Cell. Biol. 12:2967-2975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sieweke, M. H., and T. Graf. 1998. A transcription factor party during blood cell differentiation. Curr. Opin. Genet. Dev. 8:545-551. [DOI] [PubMed] [Google Scholar]
- 55.Simon, M. C. 1998. PU.1 and hematopoiesis: lessons learned from gene targeting experiments. Semin. Immunol. 10:111-118. [DOI] [PubMed] [Google Scholar]
- 56.Strauss, E. C., N. C. Andrews, D. R. Higgs, and S. H. Orkin. 1992. In vivo footprinting of the human alpha-globin locus upstream regulatory element by guanine and adenine ligation-mediated polymerase chain reaction. Mol. Cell. Biol. 12:2135-2142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Takahashi, Y., J. B. Rayman, and B. D. Dynlacht. 2000. Analysis of promoter binding by the E2F and pRB families in vivo: distinct E2F proteins mediate activation and repression. Genes Dev. 14:804-816. [PMC free article] [PubMed] [Google Scholar]
- 58.Tenen, D. G. 2003. Disruption of differentiation in human cancer: AML shows the way. Nat. Rev. Cancer 3:89-101. [DOI] [PubMed] [Google Scholar]
- 59.Thomas, D. M., S. A. Carty, D. M. Piscopo, J. S. Lee, W. F. Wang, W. C. Forrester, and P. W. Hinds. 2001. The retinoblastoma protein acts as a transcriptional coactivator required for osteogenic differentiation. Mol. Cell 8:303-316. [DOI] [PubMed] [Google Scholar]
- 60.Tsang, A. P., J. E. Visvader, C. A. Turner, Y. Fujiwara, C. Yu, M. J. Weiss, M. Crossley, and S. H. Orkin. 1997. FOG, a multitype zinc finger protein, acts as a cofactor for transcription factor GATA-1 in erythroid and megakaryocytic differentiation. Cell 90:109-119. [DOI] [PubMed] [Google Scholar]
- 61.Visvader, J. E., A. G. Elefanty, A. Strasser, and J. M. Adams. 1992. GATA-1 but not SCL induces megakaryocytic differentiation in an early myeloid line. EMBO J. 11:4557-4564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Watanabe, G., A. Howe, R. J. Lee, C. Albanese, I. W. Shu, A. N. Karnezis, L. Zon, J. Kyriakis, K. Rundell, and R. G. Pestell. 1996. Induction of cyclin D1 by simian virus 40 small tumor antigen. Proc. Natl. Acad. Sci. USA 93:12861-12866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Whyatt, D., and F. Grosveld. 2002. Cell-nonautonomous function of the retinoblastoma tumour suppressor protein: new interpretations of old phenotypes. EMBO Rep. 3:130-135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Williams, B. O., E. M. Schmitt, L. Remington, R. T. Bronson, D. M. Albert, R. A. Weinberg, and T. Jacks. 1994. Extensive contribution of Rb-deficient cells to adult chimeric mice with limited histopathological consequences. EMBO J. 13:4251-4259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yamada, T., F. Kihara-Negishi, H. Yamamoto, M. Yamamoto, Y. Hashimoto, and T. Oikawa. 1998. Reduction of DNA binding activity of the GATA-1 transcription factor in the apoptotic process induced by overexpression of PU.1 in murine erythroleukemia cells. Exp. Cell Res. 245:186-194. [DOI] [PubMed] [Google Scholar]
- 66.Yamada, T., N. Kondoh, M. Matsumoto, M. Yoshida, A. Maekawa, and T. Oikawa. 1997. Overexpression of PU.1 induces growth and differentiation inhibition and apoptotic cell death in murine erythroleukemia cells. Blood 89:1383-1393. [PubMed] [Google Scholar]
- 67.Yamamoto, H., F. Kihara-Negishi, T. Yamada, Y. Hashimoto, and T. Oikawa. 1999. Physical and functional interactions between the transcription factor PU.1 and the coactivator CBP. Oncogene 18:1495-1501. [DOI] [PubMed] [Google Scholar]
- 68.Yamamoto, M., S. Takahashi, K. Onodera, Y. Muraosa, and J. D. Engel. 1997. Upstream and downstream of erythroid transcription factor GATA-1. Genes Cells 2:107-115. [DOI] [PubMed] [Google Scholar]
- 69.Zhang, H. S., and D. C. Dean. 2001. Rb-mediated chromatin structure regulation and transcriptional repression. Oncogene 20:3134-3138. [DOI] [PubMed] [Google Scholar]
- 70.Zhang, H. S., M. Gavin, A. Dahiya, A. A. Postigo, D. Ma, R. X. Luo, J. W. Harbour, and D. C. Dean. 2000. Exit from G1 and S phase of the cell cycle is regulated by repressor complexes containing HDAC-Rb-hSWI/SNF and Rb-hSWI/SNF. Cell 101:79-89. [DOI] [PubMed] [Google Scholar]
- 71.Zhang, P., G. Behre, J. Pan, A. Iwama, N. Wara-Aswapati, H. S. Radomska, P. E. Auron, D. G. Tenen, and Z. Sun. 1999. Negative cross-talk between hematopoietic regulators: GATA proteins repress PU.1. Proc. Natl. Acad. Sci. USA 96:8705-8710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zhang, P., X. Zhang, A. Iwama, C. Yu, K. A. Smith, B. U. Mueller, S. Narravula, B. E. Torbett, S. H. Orkin, and D. G. Tenen. 2000. PU.1 inhibits GATA-1 function and erythroid differentiation by blocking GATA-1 DNA binding. Blood 96:2641-2648. [PubMed] [Google Scholar]