Abstract
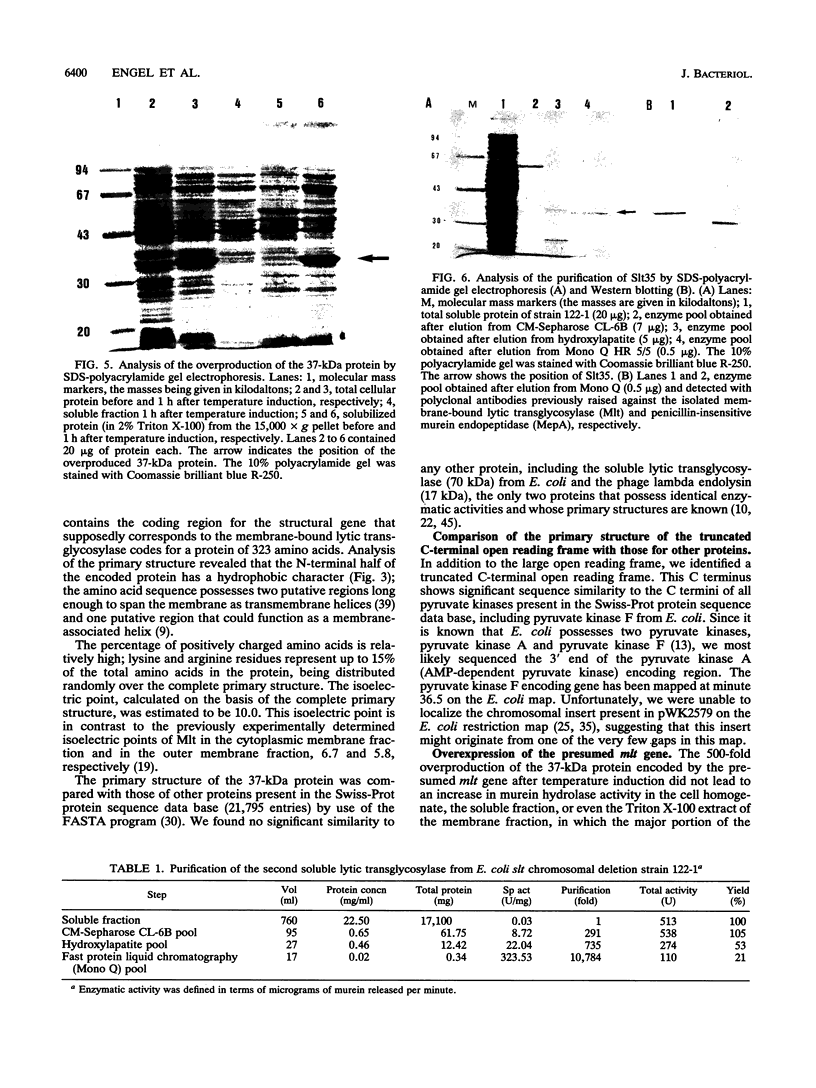
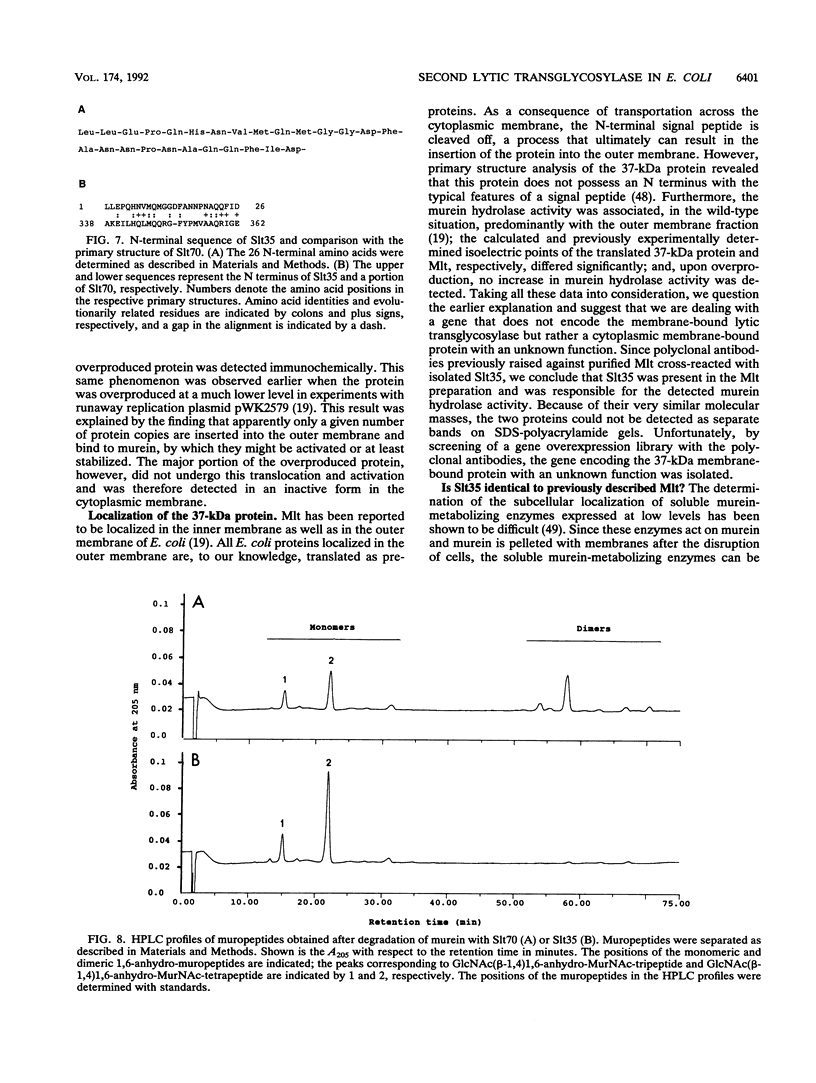
In addition to the soluble lytic transglycosylase, a murein-metabolizing enzyme with a molecular mass of 70 kDa (Slt70), Escherichia coli possesses a second lytic transglycosylase, which has been described as a membrane-bound lytic transglycosylase (Mlt; 35 kDa; EC 3.2.1.-). The mlt gene, which supposedly encodes Mlt, was cloned, and the complete nucleotide sequence was determined. The open reading frame, identified on a 1.7-kb SalI-PstI fragment, codes for a protein of 323 amino acids (M(r) = 37,410). Two transmembrane helices and one membrane-associated helix were predicted in the N-terminal half of the protein. Lysine and arginine residues represent up to 15% of the amino acids, resulting in a calculated isoelectric point of 10.0. The deduced primary structure did not show significant sequence similarity to Slt70 from E. coli. High-level expression of the presumed mlt gene was not paralleled by an increase in murein hydrolase activity. To clarify the identity of the second transglycosylase, we purified an enzyme with the specificity of a transglycosylase from an E. coli slt deletion strain. The completely soluble transglycosylase, with a molecular mass of approximately 35 kDa, was designated Slt35. Its determined 26 N-terminal amino acids showed similarity to a segment in the middle of the Slt70 primary structure. Polyclonal anti-Mlt antibodies, which had been used for the isolation of the mlt gene, were found to cross-react with Mlt as well as with Slt35, suggesting that the previously described Mlt preparation was contaminated with Slt35. We conclude that the second transglycosylase of E. coli is not a membrane-bound protein but rather is a soluble protein.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Beachey E. H., Keck W., de Pedro M. A., Schwarz U. Exoenzymatic activity of transglycosylase isolated from Escherichia coli. Eur J Biochem. 1981 May 15;116(2):355–358. doi: 10.1111/j.1432-1033.1981.tb05342.x. [DOI] [PubMed] [Google Scholar]
- Betzner A. S., Keck W. Molecular cloning, overexpression and mapping of the slt gene encoding the soluble lytic transglycosylase of Escherichia coli. Mol Gen Genet. 1989 Nov;219(3):489–491. doi: 10.1007/BF00259625. [DOI] [PubMed] [Google Scholar]
- Birnboim H. C., Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979 Nov 24;7(6):1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bittner M., Kupferer P., Morris C. F. Electrophoretic transfer of proteins and nucleic acids from slab gels to diazobenzyloxymethyl cellulose or nitrocellulose sheets. Anal Biochem. 1980 Mar 1;102(2):459–471. doi: 10.1016/0003-2697(80)90182-7. [DOI] [PubMed] [Google Scholar]
- Bolton A. E., Hunter W. M. The labelling of proteins to high specific radioactivities by conjugation to a 125I-containing acylating agent. Biochem J. 1973 Jul;133(3):529–539. doi: 10.1042/bj1330529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976 May 7;72:248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- Cookson B. T., Cho H. L., Herwaldt L. A., Goldman W. E. Biological activities and chemical composition of purified tracheal cytotoxin of Bordetella pertussis. Infect Immun. 1989 Jul;57(7):2223–2229. doi: 10.1128/iai.57.7.2223-2229.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davison J., Heusterspreute M., Chevalier N., Brunel F. A 'phase-shift' fusion system for the regulation of foreign gene expression by lambda repressor in gram-negative bacteria. Gene. 1987;60(2-3):227–235. doi: 10.1016/0378-1119(87)90231-9. [DOI] [PubMed] [Google Scholar]
- Eisenberg D., Schwarz E., Komaromy M., Wall R. Analysis of membrane and surface protein sequences with the hydrophobic moment plot. J Mol Biol. 1984 Oct 15;179(1):125–142. doi: 10.1016/0022-2836(84)90309-7. [DOI] [PubMed] [Google Scholar]
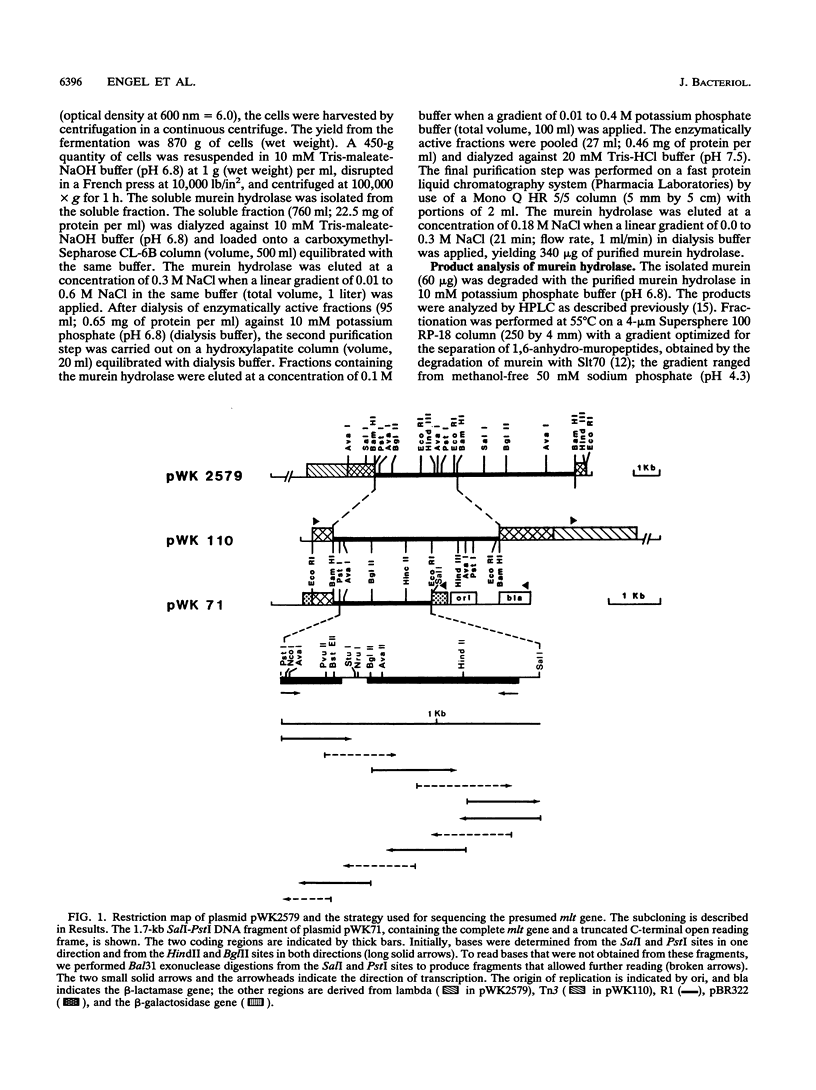
- Engel H., Kazemier B., Keck W. Murein-metabolizing enzymes from Escherichia coli: sequence analysis and controlled overexpression of the slt gene, which encodes the soluble lytic transglycosylase. J Bacteriol. 1991 Nov;173(21):6773–6782. doi: 10.1128/jb.173.21.6773-6782.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
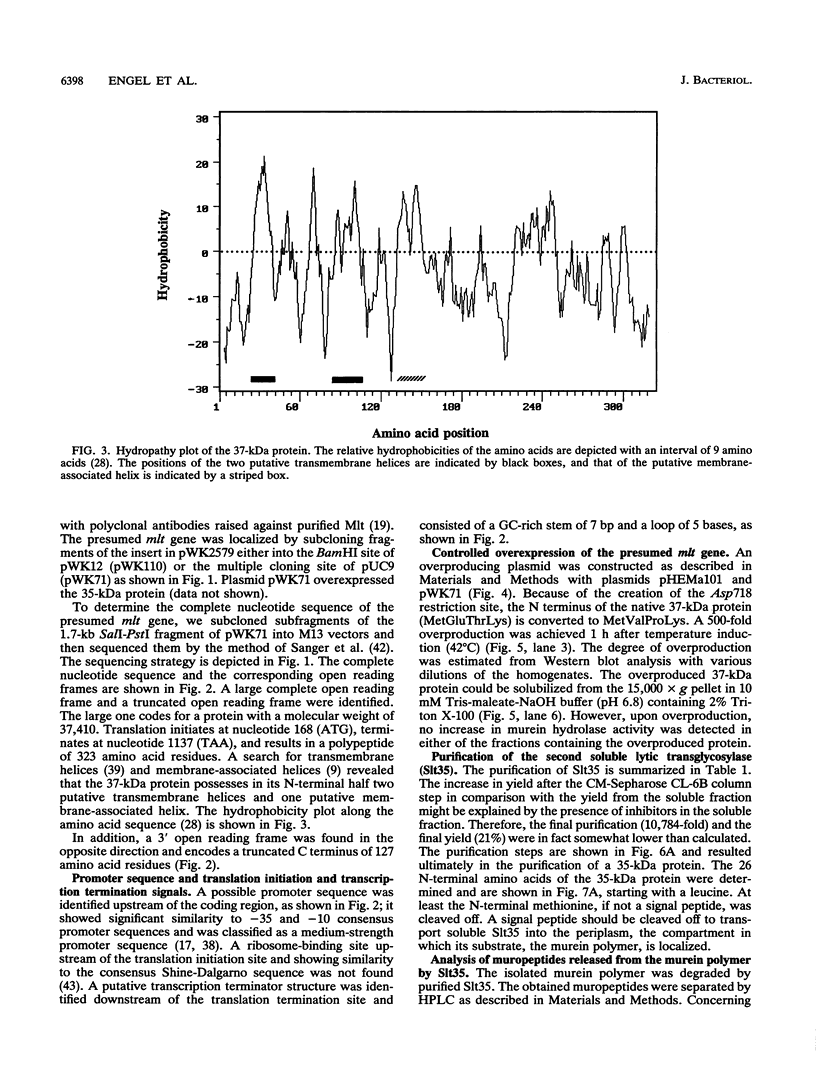
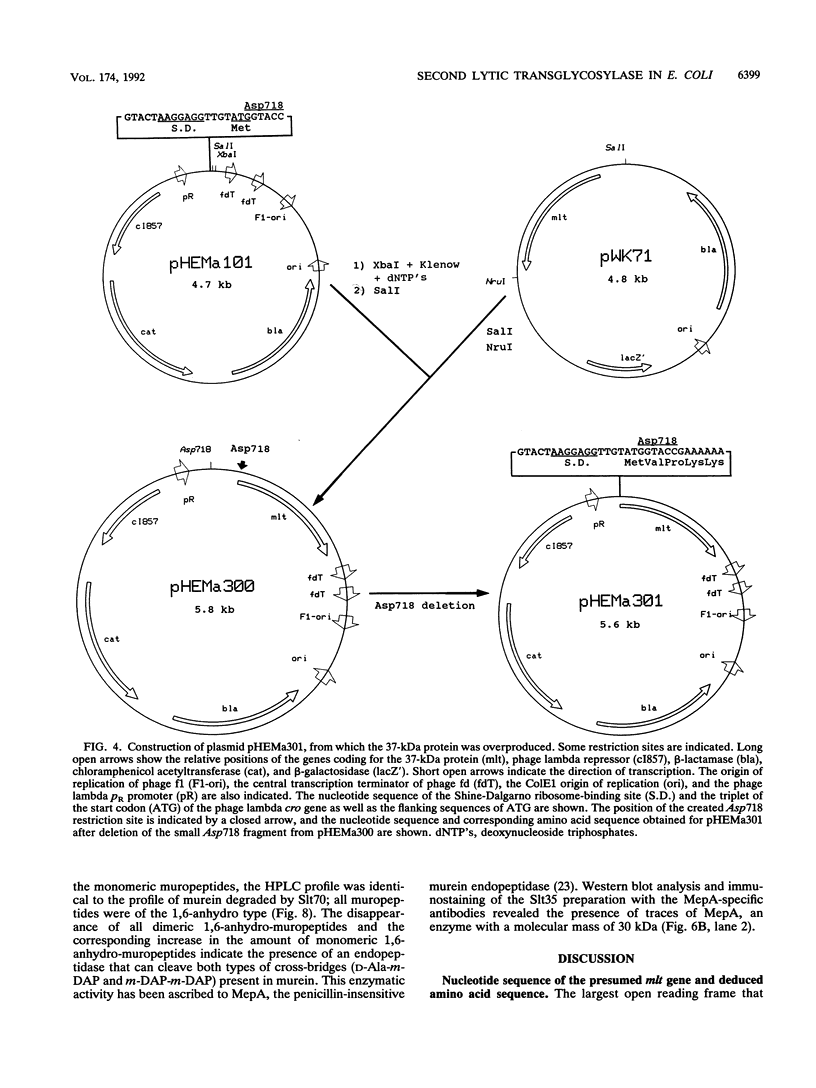
- Engel H., Mottl H., Keck W. A modified vector for the controlled high-level overproduction of staphylococcal protein A fusion proteins in the periplasm of Escherichia coli. Protein Expr Purif. 1992 Apr;3(2):108–113. [PubMed] [Google Scholar]
- Garrido-Pertierra A., Cooper R. A. Evidence for two distinct pyruvate kinase genes in Escherichia coli K-12. FEBS Lett. 1983 Oct 17;162(2):420–422. doi: 10.1016/0014-5793(83)80799-6. [DOI] [PubMed] [Google Scholar]
- Glauner B., Höltje J. V. Growth pattern of the murein sacculus of Escherichia coli. J Biol Chem. 1990 Nov 5;265(31):18988–18996. [PubMed] [Google Scholar]
- Glauner B., Höltje J. V., Schwarz U. The composition of the murein of Escherichia coli. J Biol Chem. 1988 Jul 25;263(21):10088–10095. [PubMed] [Google Scholar]
- Goodell E. W. Recycling of murein by Escherichia coli. J Bacteriol. 1985 Jul;163(1):305–310. doi: 10.1128/jb.163.1.305-310.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawley D. K., McClure W. R. Compilation and analysis of Escherichia coli promoter DNA sequences. Nucleic Acids Res. 1983 Apr 25;11(8):2237–2255. doi: 10.1093/nar/11.8.2237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helfman D. M., Feramisco J. R., Fiddes J. C., Thomas G. P., Hughes S. H. Identification of clones that encode chicken tropomyosin by direct immunological screening of a cDNA expression library. Proc Natl Acad Sci U S A. 1983 Jan;80(1):31–35. doi: 10.1073/pnas.80.1.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Höltje J. V., Tuomanen E. I. The murein hydrolases of Escherichia coli: properties, functions and impact on the course of infections in vivo. J Gen Microbiol. 1991 Mar;137(3):441–454. doi: 10.1099/00221287-137-3-441. [DOI] [PubMed] [Google Scholar]
- Karow M., Georgopoulos C. Isolation and characterization of the Escherichia coli msbB gene, a multicopy suppressor of null mutations in the high-temperature requirement gene htrB. J Bacteriol. 1992 Feb;174(3):702–710. doi: 10.1128/jb.174.3.702-710.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keck W., Wientjes F. B., Schwarz U. Comparison of two hydrolytic murein transglycosylases of Escherichia coli. Eur J Biochem. 1985 May 2;148(3):493–497. doi: 10.1111/j.1432-1033.1985.tb08866.x. [DOI] [PubMed] [Google Scholar]
- Keck W., van Leeuwen A. M., Huber M., Goodell E. W. Cloning and characterization of mepA, the structural gene of the penicillin-insensitive murein endopeptidase from Escherichia coli. Mol Microbiol. 1990 Feb;4(2):209–219. doi: 10.1111/j.1365-2958.1990.tb00588.x. [DOI] [PubMed] [Google Scholar]
- Kohara Y., Akiyama K., Isono K. The physical map of the whole E. coli chromosome: application of a new strategy for rapid analysis and sorting of a large genomic library. Cell. 1987 Jul 31;50(3):495–508. doi: 10.1016/0092-8674(87)90503-4. [DOI] [PubMed] [Google Scholar]
- Krueger J. M., Karnovsky M. L., Martin S. A., Pappenheimer J. R., Walter J., Biemann K. Peptidoglycans as promoters of slow-wave sleep. II. Somnogenic and pyrogenic activities of some naturally occurring muramyl peptides; correlations with mass spectrometric structure determination. J Biol Chem. 1984 Oct 25;259(20):12659–12662. [PubMed] [Google Scholar]
- Kunkel T. A., Roberts J. D., Zakour R. A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Methods Enzymol. 1987;154:367–382. doi: 10.1016/0076-6879(87)54085-x. [DOI] [PubMed] [Google Scholar]
- Kyte J., Doolittle R. F. A simple method for displaying the hydropathic character of a protein. J Mol Biol. 1982 May 5;157(1):105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lipman D. J., Pearson W. R. Rapid and sensitive protein similarity searches. Science. 1985 Mar 22;227(4693):1435–1441. doi: 10.1126/science.2983426. [DOI] [PubMed] [Google Scholar]
- Mandel M., Higa A. Calcium-dependent bacteriophage DNA infection. J Mol Biol. 1970 Oct 14;53(1):159–162. doi: 10.1016/0022-2836(70)90051-3. [DOI] [PubMed] [Google Scholar]
- Martin S. A., Karnovsky M. L., Krueger J. M., Pappenheimer J. R., Biemann K. Peptidoglycans as promoters of slow-wave sleep. I. Structure of the sleep-promoting factor isolated from human urine. J Biol Chem. 1984 Oct 25;259(20):12652–12658. [PubMed] [Google Scholar]
- Matsudaira P. Sequence from picomole quantities of proteins electroblotted onto polyvinylidene difluoride membranes. J Biol Chem. 1987 Jul 25;262(21):10035–10038. [PubMed] [Google Scholar]
- Melly M. A., McGee Z. A., Rosenthal R. S. Ability of monomeric peptidoglycan fragments from Neisseria gonorrhoeae to damage human fallopian-tube mucosa. J Infect Dis. 1984 Mar;149(3):378–386. doi: 10.1093/infdis/149.3.378. [DOI] [PubMed] [Google Scholar]
- Mett H., Keck W., Funk A., Schwarz U. Two different species of murein transglycosylase in Escherichia coli. J Bacteriol. 1980 Oct;144(1):45–52. doi: 10.1128/jb.144.1.45-52.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohana Rao J. K., Argos P. A conformational preference parameter to predict helices in integral membrane proteins. Biochim Biophys Acta. 1986 Jan 30;869(2):197–214. doi: 10.1016/0167-4838(86)90295-5. [DOI] [PubMed] [Google Scholar]
- Mulligan M. E., Hawley D. K., Entriken R., McClure W. R. Escherichia coli promoter sequences predict in vitro RNA polymerase selectivity. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 2):789–800. doi: 10.1093/nar/12.1part2.789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Médigue C., Hénaut A., Danchin A. Escherichia coli molecular genetic map (1000 kbp): update I. Mol Microbiol. 1990 Sep;4(9):1443–1454. [PubMed] [Google Scholar]
- Roeder W., Somerville R. L. Cloning the trpR gene. Mol Gen Genet. 1979 Nov;176(3):361–368. doi: 10.1007/BF00333098. [DOI] [PubMed] [Google Scholar]
- Rozeboom H. J., Dijkstra B. W., Engel H., Keck W. Crystallization of the soluble lytic transglycosylase from Escherichia coli K12. J Mol Biol. 1990 Apr 20;212(4):557–559. doi: 10.1016/0022-2836(90)90221-7. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shine J., Dalgarno L. The 3'-terminal sequence of Escherichia coli 16S ribosomal RNA: complementarity to nonsense triplets and ribosome binding sites. Proc Natl Acad Sci U S A. 1974 Apr;71(4):1342–1346. doi: 10.1073/pnas.71.4.1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanssens P., Opsomer C., McKeown Y. M., Kramer W., Zabeau M., Fritz H. J. Efficient oligonucleotide-directed construction of mutations in expression vectors by the gapped duplex DNA method using alternating selectable markers. Nucleic Acids Res. 1989 Jun 26;17(12):4441–4454. doi: 10.1093/nar/17.12.4441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vieira J., Messing J. The pUC plasmids, an M13mp7-derived system for insertion mutagenesis and sequencing with synthetic universal primers. Gene. 1982 Oct;19(3):259–268. doi: 10.1016/0378-1119(82)90015-4. [DOI] [PubMed] [Google Scholar]
- Walderich B., Höltje J. V. Subcellular distribution of the soluble lytic transglycosylase in Escherichia coli. J Bacteriol. 1991 Sep;173(18):5668–5676. doi: 10.1128/jb.173.18.5668-5676.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Heijne G. A new method for predicting signal sequence cleavage sites. Nucleic Acids Res. 1986 Jun 11;14(11):4683–4690. doi: 10.1093/nar/14.11.4683. [DOI] [PMC free article] [PubMed] [Google Scholar]