Abstract
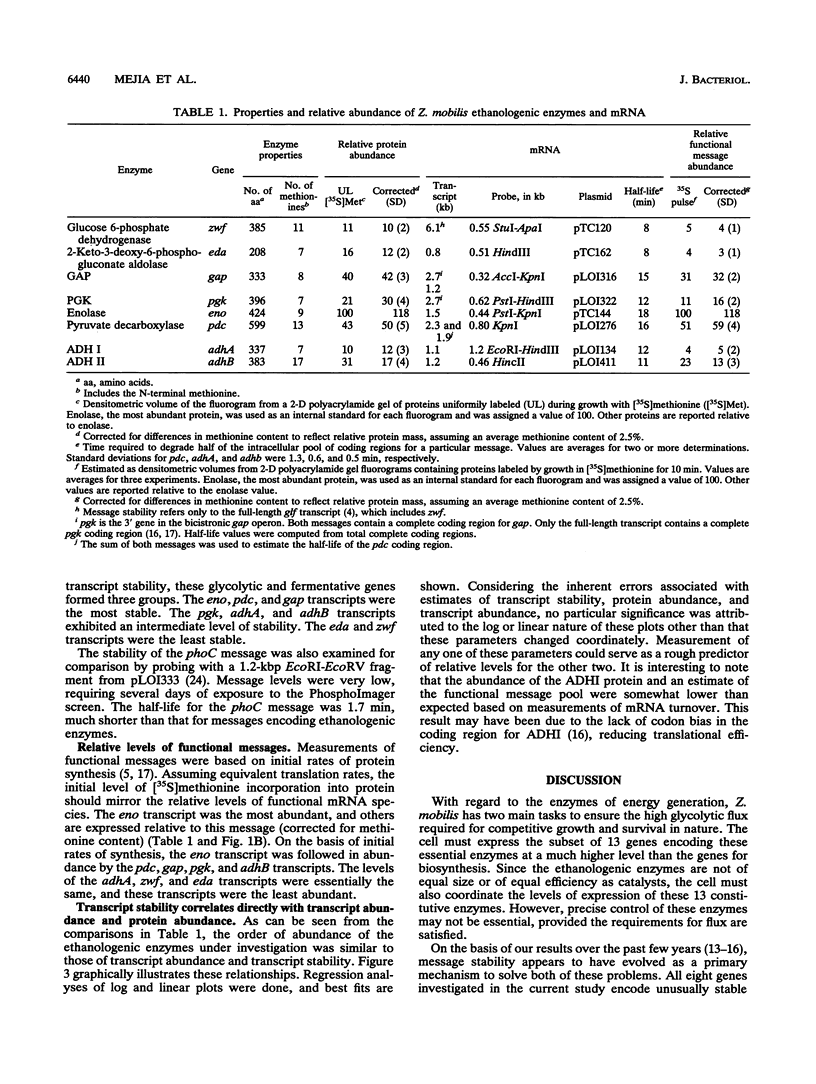
Although Zymomonas mobilis is prototrophic, glycolytic and fermentative enzymes (ethanologenic enzymes) constitute over half of the cytoplasmic protein. In this study, transcript stability, functional message pools, and the abundance of cytoplasmic products were compared for genes encoding eight of these essential enzymes. The transcripts of all were very stable, with half-lives ranging from 8 to 18 min. This transcript stability is proposed as an important feature in Z. mobilis that may distinguish highly expressed genes for energy generation from biosynthetic genes, which are required at much lower levels. The evolution of multiple promoters to enhance transcription from single-copy genes, of structural features that alter translational efficiency, and of differences in protein turnover is hypothesized to serve a subordinate role in the regulation of Z. mobilis gene expression. Among the eight ethanologenic genes examined, differences in transcript stability were found to directly correlate with differences in functional message pools and cytoplasmic protein levels. These differences in transcript stability are hypothesized to have evolved as a primary mechanism to balance the levels of individual enzymes within the glycolytic and fermentative pathways.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- An H., Scopes R. K., Rodriguez M., Keshav K. F., Ingram L. O. Gel electrophoretic analysis of Zymomonas mobilis glycolytic and fermentative enzymes: identification of alcohol dehydrogenase II as a stress protein. J Bacteriol. 1991 Oct;173(19):5975–5982. doi: 10.1128/jb.173.19.5975-5982.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnell W. O., Yi K. C., Conway T. Sequence and genetic organization of a Zymomonas mobilis gene cluster that encodes several enzymes of glucose metabolism. J Bacteriol. 1990 Dec;172(12):7227–7240. doi: 10.1128/jb.172.12.7227-7240.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belasco J. G., Higgins C. F. Mechanisms of mRNA decay in bacteria: a perspective. Gene. 1988 Dec 10;72(1-2):15–23. doi: 10.1016/0378-1119(88)90123-0. [DOI] [PubMed] [Google Scholar]
- Conway T., Fliege R., Jones-Kilpatrick D., Liu J., Barnell W. O., Egan S. E. Cloning, characterization and expression of the Zymononas mobilis eda gene that encodes 2-keto-3-deoxy-6-phosphogluconate aldolase of the Entner-Doudoroff pathway. Mol Microbiol. 1991 Dec;5(12):2901–2911. doi: 10.1111/j.1365-2958.1991.tb01850.x. [DOI] [PubMed] [Google Scholar]
- Conway T., Ingram L. O. Phosphoglycerate kinase gene from Zymomonas mobilis: cloning, sequencing, and localization within the gap operon. J Bacteriol. 1988 Apr;170(4):1926–1933. doi: 10.1128/jb.170.4.1926-1933.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conway T., Osman Y. A., Konnan J. I., Hoffmann E. M., Ingram L. O. Promoter and nucleotide sequences of the Zymomonas mobilis pyruvate decarboxylase. J Bacteriol. 1987 Mar;169(3):949–954. doi: 10.1128/jb.169.3.949-954.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conway T., Sewell G. W., Ingram L. O. Glyceraldehyde-3-phosphate dehydrogenase gene from Zymomonas mobilis: cloning, sequencing, and identification of promoter region. J Bacteriol. 1987 Dec;169(12):5653–5662. doi: 10.1128/jb.169.12.5653-5662.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conway T., Sewell G. W., Osman Y. A., Ingram L. O. Cloning and sequencing of the alcohol dehydrogenase II gene from Zymomonas mobilis. J Bacteriol. 1987 Jun;169(6):2591–2597. doi: 10.1128/jb.169.6.2591-2597.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eddy C. K., Keshav K. F., An H., Utt E. A., Mejia J. P., Ingram L. O. Segmental message stabilization as a mechanism for differential expression from the Zymomonas mobilis gap operon. J Bacteriol. 1991 Jan;173(1):245–254. doi: 10.1128/jb.173.1.245-254.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eddy C. K., Mejia J. P., Conway T., Ingram L. O. Differential expression of gap and pgk genes within the gap operon of Zymomonas mobilis. J Bacteriol. 1989 Dec;171(12):6549–6554. doi: 10.1128/jb.171.12.6549-6554.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keshav K. F., Yomano L. P., An H. J., Ingram L. O. Cloning of the Zymomonas mobilis structural gene encoding alcohol dehydrogenase I (adhA): sequence comparison and expression in Escherichia coli. J Bacteriol. 1990 May;172(5):2491–2497. doi: 10.1128/jb.172.5.2491-2497.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiley P. J., Kaplan S. Molecular genetics of photosynthetic membrane biosynthesis in Rhodobacter sphaeroides. Microbiol Rev. 1988 Mar;52(1):50–69. doi: 10.1128/mr.52.1.50-69.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neale A. D., Scopes R. K., Kelly J. M., Wettenhall R. E. The two alcohol dehydrogenases of Zymomonas mobilis. Purification by differential dye ligand chromatography, molecular characterisation and physiological roles. Eur J Biochem. 1986 Jan 2;154(1):119–124. doi: 10.1111/j.1432-1033.1986.tb09366.x. [DOI] [PubMed] [Google Scholar]
- Neale A. D., Scopes R. K., Wettenhall R. E., Hoogenraad N. J. Pyruvate decarboxylase of Zymomonas mobilis: isolation, properties, and genetic expression in Escherichia coli. J Bacteriol. 1987 Mar;169(3):1024–1028. doi: 10.1128/jb.169.3.1024-1028.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Farrell P. H. High resolution two-dimensional electrophoresis of proteins. J Biol Chem. 1975 May 25;250(10):4007–4021. [PMC free article] [PubMed] [Google Scholar]
- Osman Y. A., Conway T., Bonetti S. J., Ingram L. O. Glycolytic flux in Zymomonas mobilis: enzyme and metabolite levels during batch fermentation. J Bacteriol. 1987 Aug;169(8):3726–3736. doi: 10.1128/jb.169.8.3726-3736.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pawluk A., Scopes R. K., Griffiths-Smith K. Isolation and properties of the glycolytic enzymes from Zymomonas mobilis. The five enzymes from glyceraldehyde-3-phosphate dehydrogenase through to pyruvate kinase. Biochem J. 1986 Aug 15;238(1):275–281. doi: 10.1042/bj2380275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pond J. L., Eddy C. K., Mackenzie K. F., Conway T., Borecky D. J., Ingram L. O. Cloning, sequencing, and characterization of the principal acid phosphatase, the phoC+ product, from Zymomonas mobilis. J Bacteriol. 1989 Feb;171(2):767–774. doi: 10.1128/jb.171.2.767-774.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scopes R. K., Griffiths-Smith K. Use of differential dye-ligand chromatography with affinity elution for enzyme purification: 6-phosphogluconate dehydratase from Zymomonas mobilis. Anal Biochem. 1984 Feb;136(2):530–534. doi: 10.1016/0003-2697(84)90257-4. [DOI] [PubMed] [Google Scholar]
- Scopes R. K., Testolin V., Stoter A., Griffiths-Smith K., Algar E. M. Simultaneous purification and characterization of glucokinase, fructokinase and glucose-6-phosphate dehydrogenase from Zymomonas mobilis. Biochem J. 1985 Jun 15;228(3):627–634. doi: 10.1042/bj2280627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scopes R. K. Use of differential dye-ligand chromatography with affinity elution for enzyme purification: 2-keto-3-deoxy-6-phosphogluconate aldolase from Zymomonas mobilis. Anal Biochem. 1984 Feb;136(2):525–529. doi: 10.1016/0003-2697(84)90256-2. [DOI] [PubMed] [Google Scholar]




