Abstract
The uninduced Drosophila hsp70 gene is poised for rapid activation. Here we examine the rapid changes upon heat shock in levels and location of heat shock factor (HSF), RNA polymerase II (Pol II) and its phosphorylated forms, and the Pol II kinase P-TEFb on hsp70 in vivo by using both real-time PCR assays of chromatin immunoprecipitates and polytene chromosome immunofluorescence. These studies capture Pol II recruitment and progression along hsp70 and reveal distinct spatial and temporal patterns of serine 2 and serine 5 phosphorylation: in uninduced cells, the promoter-paused Pol II shows Ser5 but not Ser2 phosphorylation, and in induced cells the relative level of Ser2-P Pol II is lower at the promoter than at regions downstream. An early time point of heat shock activation captures unphosphorylated Pol II recruited to the promoter prior to P-TEFb, and during the first wave of transcription Pol II and the P-TEFb kinase can be seen tracking together across hsp70 with indistinguishable kinetics. Pol II distributions on several other genes with paused Pol II show a pattern of Ser5 and Ser2 phosphorylation similar to that of hsp70. These studies of factor choreography set important limits in modeling transcription regulatory mechanisms.
Recent studies of posttranslational modifications of, and factor interactions with, the C-terminal domain (CTD) of the largest subunit of RNA polymerase II (Pol II) have implicated this region of Pol II in productive transcriptional elongation and the coupling of transcription and pre-mRNA processing (for reviews see references 32 and 37). The CTD is highly conserved among eukaryotes, containing multiple repeats of a consensus heptad YSPTSPS; mammalian Pol II CTD consists of 52 repeats, yeast consists of 25 to 26 repeats, while Drosophila has 42 repeats (1, 6). This large flexible arm of Pol II appears to serve as a docking site for, or to stimulate the recruitment of, an orchestrated assembly of factors involved in pre-mRNA capping, splicing, and 3′ polyadenylation at different stages in production of the nascent transcript (5, 9, 13, 14, 20, 26). One way this coordination has been proposed to occur is through the known phosphorylation of serines 2 and 5 (Ser2-P and Ser5-P, respectively) of the heptad repeat (52, 56). Cdk7, the kinase subunit of general transcription factor TFIIH, has been shown to phosphorylate the CTD at Ser5, an event proposed to occur early in the transcription cycle (12, 24, 57). This in turn appears to influence the association and activity of the capping machinery (5, 15, 18, 27, 28, 42, 45). Positive transcription elongation factor b (P-TEFb) is able to phosphorylate the CTD at Ser2 and, under certain conditions, Ser5 (25, 38, 57). P-TEFb is a kinase composed of the proteins cyclin T (CycT) and cdk9 and is known to be recruited upon gene activation, overcoming the negative effects of factors like Spt5 and negative elongation factor (50) and aiding in the transition from transcription initiation to elongation (36). Cdk8, a component of the coactivator complex Mediator, has also been shown to phosphorylate Ser5 in vitro, although its role in transcription regulation is not yet fully understood (12, 16, 17, 41).
While the broadly accepted model of gene activation centers on Pol II recruitment upon activator binding to the promoter, an interesting divergence from this model stems from the initial studies of the hsp70 gene in Drosophila melanogaster. Here the uninduced state maintains a paused, transcriptionally engaged Pol II molecule immediately downstream of the transcription start site (39, 44). Also present prior to gene activation are the promoter-binding protein GAGA factor (29), TATA-binding protein (10), and Spt5 (2). The promoter maintains an open configuration (7, 53), and mRNA transcripts associated with the paused Pol II molecule undergo 5′ mRNA capping as Pol II moves through the pause region (40). Immunofluorescence analysis of polytene chromosomes that have undergone heat shock reveals rapid recruitment of the activator heat shock factor (HSF) as well as P-TEFb, Spt6, Mediator, and additional Spt5 and Pol II (2, 21, 33). Artificial recruitment of P-TEFb to hsp70 is sufficient to induce transcription in the absence of heat shock, implicating it as a major player in overcoming a rate-limiting step at the early stages of Pol II's maturation into an elongation complex (21).
Using general antibodies to hypophosphorylated Pol II (Pol IIa) and hyperphosphorylated Pol II (Pol IIo), previous work has shown that the paused Pol II on hsp70 is hypophosphorylated and that phosphorylation increases upon heat shock-induced transcriptional activation (30, 51). Recent studies in other systems have analyzed the Pol IIo forms by using antibodies directed specifically toward Ser2 or Ser5 phosphorylation (4, 35). Komarnitsky et al. employed chromatin immunoprecipitation (ChIP) assays in yeast to analyze the phosphorylation status of the CTD at the promoter compared to that of regions in the open reading frame (ORF) on TFIIH-dependent genes (18). While Pol II distribution appeared to remain constant along the gene, Ser5-P signal was much stronger at the promoter than at the ORF. Conversely, Ser2-P was only detected in the ORF. Soutoglou and Talianidis saw a similar pattern of Ser5-P distribution on the alpha-AT gene in human enterocytes but detected Ser2-P at both the promoter and downstream regions (48). These changes in phosphorylation patterns on the CTD may distinguish the Pol II machinery as it progresses through the gene, signaling distinct transcriptional processes and coordinated RNA processing events.
By examining a gene that can be abruptly and synchronously activated in cells and by documenting precisely when and where on this gene the regulatory factors and the transcription machine (and its modified states) interact, we have been able to tease out several insights into hsp70 gene activation. ChIP assays were used to track changes in the protein densities of HSF, Pol II, and P-TEFb and in Pol II phosphorylation status in the seconds and minutes following a synchronous and robust heat shock induction. Further, real-time PCR analysis of this ChIP material was sensitive and quantitative and was easily scaled up to allow repeated analysis of multiple time points, factors, and DNA regions. A kinetic analysis of changes in immunofluorescence on polytene chromosomes at a transgenic locus containing a single hsp70-lacZ gene provided independent corroboration of the patterns of factor recruitment obtained by ChIP. Genes previously shown by run-on analysis to harbor a paused polymerase, hsp26, beta-1-tubulin (Tub), glyceraldehyde-3-phosphate dehydrogenase (GAP), and Actin5C, were analyzed along with a control gene, histone H1 (43), for Pol II modification states to evaluate whether promoter-associated Pol II pausing and Pol II phosphorylation patterns correlate. Taken together, this approach defines the temporal and spatial features of specific regulatory models.
MATERIALS AND METHODS
ChIP.
Extraction of cross-linked protein-DNA complexes was performed as previously described, with some adjustments (33). Briefly, Kc cells were grown to 5 × 106 to 7 × 106 cells/ml in serum-free HyQ-CCM3 medium (HyClone Laboratories, Inc.). Cells for heat shock time points were subjected to instantaneous heat shock by addition of an equivalent volume of medium preheated to 48°C to the growing cells, which were at 25°C. After holding the cells at the heat shock temperature, 36.5°C, for the allotted interval the cells were immediately cooled down to the temperature of control, non-heat shocked cells with the addition of 1/3 total volume of 4°C medium immediately prior to cross-linking. This cooling to room temperature allows direct comparison of heat shocked and control cells, avoiding the increased efficiency of formaldehyde cross-linking at the higher temperature over that at the control temperature. Cells were cross-linked with a 0.3% formaldehyde final concentration solution (3.3% formaldehyde, 0.1 M NaCl, 1 mM EDTA, 0.5 mM EGTA, 50 mM Tris [pH 8.0]) for 1 min. Quenching of cross-linking by 125 mM final glycine was then performed. Cells were spun down and resuspended in sonication buffer (10 mM Tris [pH 8.0], 1 mM EDTA, 0.5 mM EGTA, 0.5 mM phenylmethylsulfonyl fluoride) with protease inhibitor cocktail (Roche). Cells were sonicated with a Sonifier 350 (Branson Sonic Power Co.) for 20-s bursts 40 times on ice. DNA fragments, on average, were less than 700 bp as noted by agarose gel analysis and HSF control immunoprecipitations. After spinning down debris the supernatant was mixed with an equivalent volume of 6 M urea and was dialyzed overnight (10 mM Tris [pH 8.0], 1 mM EDTA, 0.5 mM EGTA, 0.5 mM phenylmethylsulfonyl fluoride, 10% glycerol, 0.1% sodium deoxycholate, 1.0% Triton-X 100). Extracts were then spun again and aliquoted for storage at −80°C.
Immunoprecipitations were performed as described in the Santa Cruz Biotechnology ChIP assay kit, with minor adjustments, including six low-salt washes and three high-salt washes. A total of 2.5 × 107 cells of each extract were used per immunoprecipitation. The amount of antibody per immunoprecipitation was as follows: HSF, 1 μl (2); 8WG16, 1 μl (Covance); Pol II (total), 5 μl (lab stock purified by Ed Wang); H5, 6 μl (Covance); H14, 3 μl (Covance); and P-TEFb, 2 μl (affinity purified CycT kindly provided by David Price) (21). Immunoprecipitations were resuspended in 50 μl of 10 mM Tris (pH 8.0) for PCR analysis.
Real-time PCR analysis.
Real-time PCR was performed by using 2 μl of each ChIP sample (106 cells). A volume of 7.5 μl of SYBR Green PCR Master Mix (Applied Biosystems), 1 μl of 5 μM forward and reverse primers, and 3.5 μl of H2O were added to each sample for analysis by absolute quantification. A 5-point serial dilution of each extract was run concurrently with each extract for every gene region tested. The primers were designed by using the Primer Express software (Applied Biosystems) and are as follows: hsp70, −200F 5′-TGCCAGAAAGAAAACTCGAGAAA; hsp70, −108R 5′-GACAGAGTGAGAGAGCAATAGTACAGAGA; hsp70 + 4F, 5′-CAATTCAAACAAGCAAAGTGAACAC; hsp70 + 112R, 5′-TGATTCACTTTAACTTGCACTTTA; hsp70 + 334F, 5′-CACCACGCCGTCCTACGT; hsp70 + 423R, 5′-GGTTCATGGCCACCTGGTT; hsp70 + 645F, 5′-ATATCTGGGCGAGAGCATCACA; hsp70 + 718R, 5′-GTAGCCTGGCGCTGGGAGTC; hsp70 + 872F, 5′-CATCGACGAGGGATCTCTGTTC; hsp70 + 1019R, 5′-GGCGCGAGGGTTGGA; hsp70 + 1363F, 5′-CTGTGCAGGCCGCTATCC; hsp70 + 1490R, 5′-GCGCTCGATCAGCTTGGT; hsp70 + 1649F, 5′-GGGTGTGCCCCAGATAGAAG; hsp70 + 1754R, 5′-TGTCGTTCTTGATCGTGATGTTC; hsp26-22F, 5′-CGAACAGAGCACAGATCGAATTC; hsp26 + 63R, 5′-TTTCTCGTTTGAAGTTTTGCTTTTG; hsp26 + 580F, 5′-CAAGGTTCCCGATGGCTACA; hsp26 + 667R, 5′-CTGCGGCTTGGGAATACTGA; Tub-70F, 5′-CACTAAAACGGGATTTGCGTTT; Tub + 11R, 5′-GGAGAGCGCGGGCTTT; Tub + 2075F, 5′-GGCGGAACGCAATGACA; Tub + 2152R, 5′-CGCCACACCAACGATAACG; Actin5C-3F, 5′-GAGTTCTTGTGCTGTGTGGATACTC; Actin5C + 90R, 5′-TTAAGTCTTTCGGTTTGGTGTCTCT; Actin5C + 1781F, 5′-GGAAATCCGCATTCTTTCCA; Actin5C + 1848R, 5′-CGACAACCAGAGCAGCAACTT; H1-64F, 5′-TGTTAAAGTGCTCTCCTCCTCGAT; H1-5R, 5′-GCGCGCTGCCTACCAA; H1 + 329F, 5′-CGTGGTCAATGGAAAGCTTATTC; and H1 + 397R, 5′-GAGGCCGACAGTTTGAAAGATC.
Real-time PCR was performed in 384-well plates with the ABPrism 7900 Sequence Detection System; data was analyzed with SDS software v2.0 (Applied Biosystems). The results of real-time PCR are expressed as a point in the linear range of amplification for each sample, termed the Ct value. Percentage input for each data point was determined by comparing the Ct value of each sample to a standard curve generated from a 5-point serial dilution of genomic input (chromatin-extracted material that did not undergo immunoprecipitation). Each immunoprecipitation was performed and assayed at least four times from at least two separate protein cross-linked DNA extracts for each time point. The percent input shown for each point has a no antibody control value subtracted from the sample value to account for general background. Supplemental data can be viewed at http://www.mbg.cornell.edu/lis/online.html.
Indirect immunofluorescence.
Polytene squashes were performed as previously described with minor modifications to the heat shock procedure (21). Briefly, heat shocked larvae were placed near a small piece of moist filter paper on a piece of Saran Wrap, which was then sealed shut. The Saran Wrap was then submerged in a 36.5°C water bath for an appropriate time. Squashes were performed immediately thereafter, as was immunofluorescence staining. Briefly, slides were washed in Tris-buffered saline (TBS) and were blocked for 15 min in TBS with 5% donkey serum (Jackson Laboratories, Inc.). Antibodies were added, in TBS with 5% donkey serum, at the following dilutions: HSF, 1:100 (lab stock); H5, 1:50 (Covance, Inc.); H14, 1:50, (Covance, Inc.); and CycT, 1:50 (kindly provided by David Price). Chromosomes were incubated with primary antibodies overnight at 4°C, and then the slides were washed in TBS and were treated with the appropriate secondary antibody at 1:500 (Jackson Laboratory, Inc., or Molecular Probes, Inc.) in TBS with donkey serum for 1 h at 25°C. Slides were then washed in TBS, stained with Hoechst 33258 (Sigma), mounted in antifade solution, and analyzed. Analysis was performed by using a Zeiss Axioplan 2 microscope and OpenLab 3.0.7 imaging software. Images were taken by using the OpenLab z-series capture sequence and three-dimensional restoration.
RESULTS
Kinetic analysis of hsp70 activation.
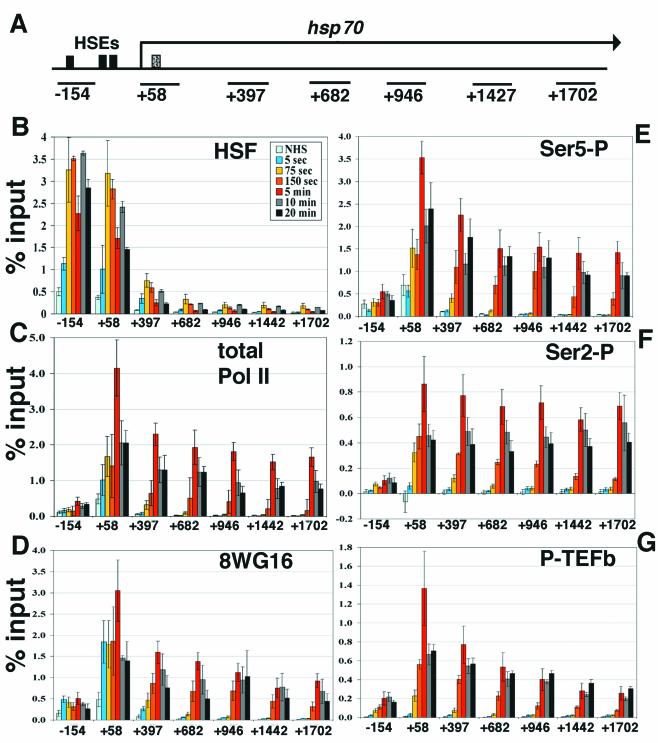
We performed ChIP analysis of Kc cell extracts taken at several time points following activation by heat shock to map the temporal and spatial recruitment, movement, and modification of factors known to be involved in hsp70 transcription. The heat shock induction was instantaneous: cells in suspension and warm media were mixed to a final temperature of 36.5°C and were maintained at this temperature. The formaldehyde cross-linking was likewise as rapid as possible: formaldehyde cross-linking times were reduced to 1 min to provide a snapshot of what was occurring during the early points of transcriptional activation. Because formaldehyde reactivity increases with temperature, following heat shock cells were brought back to room temperature with cold media immediately before the 1-min formaldehyde treatment. High spatial resolution was achieved by designing primer sets for real-time PCR to several regions along the hsp70 gene (Fig. 1A). An upstream primer set, centered at −154, detects the heat shock elements (HSE) within and adjacent to this region (54), the +58 primer set contains the region of the paused polymerase, and five others lie downstream within the ORF extending towards the end of the gene (1,925 bp). Real-time PCR analysis was performed in 384-well plates, allowing for all hsp70 gene regions and multiple proteins for various time points to be analyzed simultaneously. Duplicate samples analyzed by real-time PCR for each immunoprecipitation were generally within 1%. The majority of the data presented here express the results as a percentage of the amount of genomic material input in each immunoprecipitation. The standard error reported with each measurement is the variation associated with multiple independent ChIP assays. Using this approach, we examined activator recruitment, Pol II recruitment, elongation, and phosphorylation status as well as recruitment of the CTD kinase P-TEFb. The results from these assays are summarized in Fig. 1B to G.
FIG. 1.
Kinetic analyses of hsp70 transcription by ChIP and real-time PCR. (A) Schematic of hsp70. Solid black boxes indicate HSE sites (−175, −75, −55 from transcription start site) (54), and the dotted box indicates the site of paused Pol II (+21 to +35) (39). Black lines below the gene indicate the regions amplified by primer sets (see Materials and Methods), identified by the nucleotide at the center of the amplified region. (B) Time course of HSF recruitment on hsp70. ChIP samples were taken at several time points, analyzed by real-time PCR with the primer sets along hsp70 shown in panel A, and compared to percentage input material. Samples shown are NHS and 5-s, 75-s, 150-s, 5-min, 10-min, and 20-min instantaneous heat shock. The x axis is the hsp70 gene in nucleotides, and the y axis is percentage input. The identity of each time point line is located in the upper right-hand corner of the graph. Standard error bars are shown; n = 4, 3, 7, 4, 4, 5, and 4, respectively. (C) Kinetics of total Pol II. Standard error bars are shown; n = 4, 4, 8, 3, 4, 4, and 4, respectively. (D) Antibody 8WG16, which detects unphosphorylated Pol II CTD repeats. Standard error bars are shown; n = 4, 4, 7, 4, 5, 4, and 4, respectively. (E) Time course of Ser5 phosphorylation, detected by antibody H14. Standard error bars are shown; n = 4, 3, 7, 4, 5, 4, and 4, respectively. (F) Ser2 phosphorylation kinetics on hsp70, detected by H5. Standard error bars are shown; n = 4, 4, 8, 3, 4, 4, and 4, respectively. (G) ChIP analysis of P-TEFb by using an antibody to CycT. Standard error bars are shown; n = 3, 3, 8, 4, 5, 4, and 4, respectively.
HSF levels are saturating on the hsp70 promoter within 75 s of heat shock.
HSF is the critical activator of heat shock genes and is known to be recruited to heat shock loci. Low levels of HSF associate with the hsp70 promoter prior to heat shock (non-heat shock [NHS]) (Fig. 1B), as has been seen previously (2). HSF levels increase significantly at the promoter in as little as 5 s after heat shock. HSF levels at the promoter reach saturating levels by 75 s, remaining essentially constant through the 20-min time course. HSF is at high density on DNA detected by primer set −154, which covers known HSEs; HSF is also detected at the +58 region, which is near the HSE element at −55, and would be easily detected by the +58 probe given the average size of the sonicated chromatin used in ChIP assays. HSF signal drops off an average of fivefold from the +58 region to the +379 region, indicating the assay has the spatial resolution to distinguish the promoter from the beginning of the ORF. HSF is essentially absent from the body and 3′ end of the gene, as would be expected, since no HSEs reside in these regions.
The initial RNA Pol II molecule elongates rapidly into and through hsp70.
Pol II recruitment and its movement into the gene rapidly follows the increase in HSF. To detect the rapidity with which Pol II is recruited to and elongates through hsp70, total Pol II levels on hsp70 before and during heat shock were assayed by using an antibody to RpII215, the largest subunit of Pol II. This antibody detects all forms of Pol II, indiscriminate of the phosphorylation status of the CTD. As expected, RNA Pol II is detected at the 5′ coding region in uninduced cells (Fig. 1C) (2, 11). Pol II recruitment to the +58 region is evident at 5 s and elongates into the beginning of the gene by 75 s. At 150 s, Pol II levels at the promoter appear similar to that at 75 s, suggesting that this time period may encompass the first wave of transcriptional response. At this time point, Pol II can be detected through to the 3′ end of the gene. These data correlate well with the known elongation rate on the hsp70 gene, previously determined to be approximately 1.2 kb/min (31). The maximum Pol II signal occurs at 5 min, decreasing 30 to 50% over the entire gene by 10 and 20 min. This result is in agreement with previously reported rates of hsp70 RNA synthesis, which peaked at 5 min (49). At all time points tested where Pol II has proceeded through the gene, Pol II signals remain stronger at the start site compared to that at the ORF (approximately 2.5-fold), consistent with the observation that escape of promoter-paused Pol II remains rate-limiting even during heat shock (10). As a control, Pol II was tested at all time points on the ORF of histone H2B, a gene whose transcription is not altered by heat shock (11), and no significant changes were noted (data not shown).
The spatial resolution afforded by these assays can be seen by comparing HSF and Pol II signals along hsp70 at 5 min following heat shock (Fig. 1B and C). HSF signals are high on the two upstream fragments and drop off dramatically in the ORF. In contrast, Pol II is detected throughout the gene but is barely detected on the upstream −154 fragment.
The dynamics of Pol II CTD phosphorylation along hsp70 before and during heat shock.
Pol II can exist in multiple phosphorylation states before and during transcription, and these states can be assayed by a battery of antibodies to specific phosphorylated residues of the CTD. Kinetic analysis of hsp70 transcription using the antibody 8WG16, which recognizes hypophosphorylated CTD, shows that Pol II is present near the start site of transcription prior to heat shock, in agreement with previous work showing that promoter-paused Pol II is hypophosphorylated (Fig. 1D) (30). The 8WG16 distribution is similar to that of total Pol II in that its concentration at the promoter is greater than that at the ORF for all time points tested, and the relative signals throughout the gene were comparable. It is important to note that 8WG16 can still recognize Pol II molecules that have some level of phosphorylation, as the YSPTSPT repeats along the CTD are not all necessarily phosphorylated or unphosphorylated (30).
Figure 1E and F shows the changes in the phosphorylated CTD (Pol IIo) distribution on hsp70 during transcription. Specific antibodies H14 and H5 preferentially detect phosphorylation at Ser5 and Ser2, respectively. Interestingly, Ser5-P is detected at the start site of transcription under NHS conditions, albeit at a modest level, indicating that the paused, transcriptionally engaged polymerase contains some CTD repeats that are phosphorylated at this residue. Upon heat shock this phosphorylation increases at the promoter (+58) by 75 s and peaks at 5 min. The Ser5-P-modified Pol II first appears on the downstream fragment (+379) at 75 s and first appears at the end of the gene by 150 s. The levels of Ser5-P Pol II reach a maximum over the entire gene at 5 min. In all time points tested where Pol II has transcribed completely through the gene, Ser5-P Pol II appears to be 2.5- to 3-fold greater at the transcription start site than at the ORF. Ser2-P-modified Pol II, unlike Ser5-P, is not detected prior to heat shock but increases at the start site of transcription and enters the downstream fragment by 75 s (Fig. 1F). The Ser2-P Pol II, like the other Pol II forms, is detected at the 3′ end of the gene by 150 s and reaches a maximum over the entire gene after 5 min. Unlike other Pol II forms, the Ser2-P modification is at a uniform level throughout the gene during active elongation.
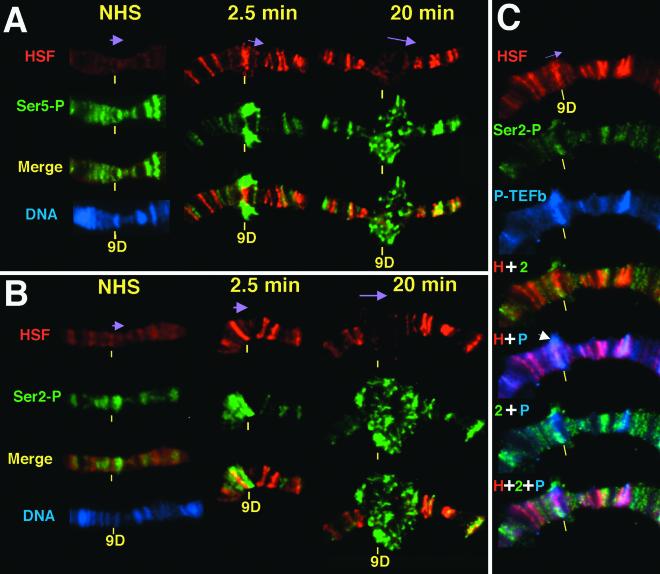
Antibody staining of polytene chromosomes provides an independent method for assessing the recruitment of proteins on heat shock genes during gene activation. Moreover, a single-copy transgenic line that contains an hsp70-lacZ fusion gene at the 9D locus and the large puffs that are generated when this gene is activated provide sufficient resolution to allow relative distribution in the promoter proximal and distal regions to be resolved (23, 47). Figure 2 shows that promoter-associated HSF resolves from transcribing Pol II at the 9D locus. At 2.5 min following heat shock of third-instar larvae, HSF levels at this site increase relative to the faint band seen on uninduced larval chromosomes (Fig. 2A). The puff at this time is still in the early stages of forming, and the HSF staining appears as a distinct band. The anti-HSF and anti-Ser5-P staining at 9D appear in some spreads as overlapping but distinct bands (data not shown) and in other spreads as a lateral decondensation that circumscribes the chromosome, which in the flattened chromosome like that shown in Fig. 2A can appear as bulges on both sides of the chromosomal puff site. By 20 min the HSF signal in the large puff had become more diffuse, presumably as a result of the large amount of chromatin decondensation in and around the activated gene (47). The Ser5-P staining was still more extensive, often creating a large halo at the outer regions of the puff encircling the nonoverlapping HSF signal at the core. The lack of overlap of the HSF and Pol II Ser5-P stainings was most evident in optical sections of such puffs (see supplemental data for a movie showing the staining pattern in three dimensions at http://www.mbg.cornell.edu/lis/online.html).
FIG. 2.
Kinetics of Pol II phosphorylation during heat shock induction on Drosophila polytene chromosomes and localization of HSF, Ser2-P, and P-TEFb in the active hsp70 puff. Shown is the transgenic hsp70-lacZ gene, located at 9DE in Bg9 flies (23), precisely mapped by electron microscopy to the end of 9D (46). Chromosomes were prepared as previously described (21) and were stained with antibodies to HSF (red) and either Ser5-P (green) in panel A or Ser2-P (green) in panels B and C, as well as P-TEFb (blue) in panel C. Purple arrows indicate the direction of transcription. (A and B) Time course is as indicated. Images underwent three-dimensional restoration. (C) Chromosomes were heat shocked for 2.5 min. Merged images show the various combinations of HSF (H), Ser2-P (2), and P-TEFb (P) immunofluorescence, as indicated by the plus sign. The white arrow indicates where P-TEFb and Ser2-P expand into the puff, resolving from HSF. These images did not undergo three-dimensional restoration.
The pattern of Ser2 phosphorylation detected by ChIP is also apparent by polytene chromosome immunofluorescence, which was performed at different time points during hsp70 gene activation (Fig. 2B). Ser2 phosphorylation is absent prior to heat shock as indicated by the low background at the transgenic site, which is reproducibly observed (Fig. 2B, NHS). Note that in the case of anti-Ser5-P staining of uninduced chromosomes, a weak band that coincides with the transgenic site is seen reproducibly (Fig. 2A). This pair of findings is consistent with the modest but significant level of Ser5-P and the absence of Ser2-P at the promoter proximal fragment as detected by ChIP. After heat shock the Ser2-P can be seen to resolve from HSF and, like Ser5-P, is spread into the outer periphery of the puff. Therefore, these independent cytological views are in complete agreement with the higher resolution ChIP assays.
P-TEFb tracks with the first wave of Pol II through hsp70.
In considering the mechanism by which Pol II matures into a phosphorylated elongation complex, the kinases implicated in this phosphorylation are of particular interest. P-TEFb kinase was analyzed through the time course with an antibody to one of its subunits, CycT (Fig. 1G). P-TEFb is not detected prior to heat shock, in agreement with previous reports (2, 21). P-TEFb is apparent at the hsp70 promoter at 75 s of heat shock, concomitant with the appearance of Ser2-P, and P-TEFb travels through the gene at a rate indistinguishable from that of total Pol II. The peak of P-TEFb signal at 5 min agrees well with a time course of P-TEFb recruitment performed by immunofluorescence on polytene chromosomes, which reached a plateau at 5 min for up to 1 h of heat shock (21).
As P-TEFb has been implicated in phosphorylation of the CTD at Ser2, polytene chromosome tricolor immunofluorescence was performed with HSF, Ser2-P, and P-TEFb at a short heat shock time point of 2.5 min, which is when a transcription puff is already forming (Fig. 2C). At a transgenic locus we observe that HSF and Ser2-P Pol II begin to resolve (as also seen by comparing Fig. 1B and F), with Pol II forming a halo around the more centrally localized HSF. Moreover, P-TEFb also resolves from HSF, indicating that it too is not restricted to the promoter. The patterns of P-TEFb and Pol II staining are very similar though not identical, in that P-TEFb shows more overlap with promoter-bound HSF than does Pol II Ser2-P.
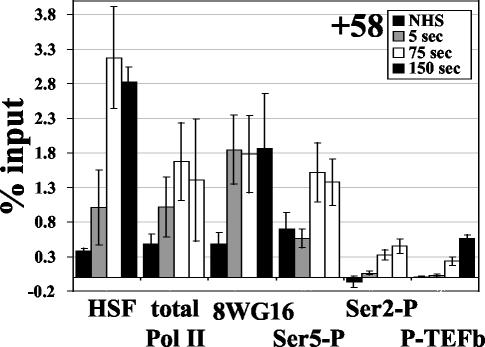
An early snapshot of gene activation: Pol II recruitment prior to phosphorylation.
The earliest heat shock time point of 5 s reveals an interesting dissection of the early events of hsp70 gene activation (Fig. 3). A moderate level of HSF recruitment to the promoter is apparent at this time point, and an increase in total Pol II can also be detected at +58. Interestingly, an increase in Pol IIa is detected at the promoter, but no significant change in Ser5-P or Ser2-P levels are apparent, suggesting that at this extremely early time we have captured the recruitment of unphosphorylated Pol II to the hsp70 promoter. While this time point may not strictly be 5 s due to the 1-min cross-linking time, it is certainly no more than 65 s after gene induction. The undetectable recruitment of P-TEFb (CycT) at this 5-s time point is consistent with the lack of phosphorylation; thus, P-TEFb is associating with the promoter after the 5-s time point, and detection of P-TEFb is concomitant with phosphorylation of the CTD.
FIG. 3.
Early events of transcription at the hsp70 promoter. Comparison of recruitment kinetics of factors or modifications at the earliest time points of transcription at the region encompassing the paused Pol II.
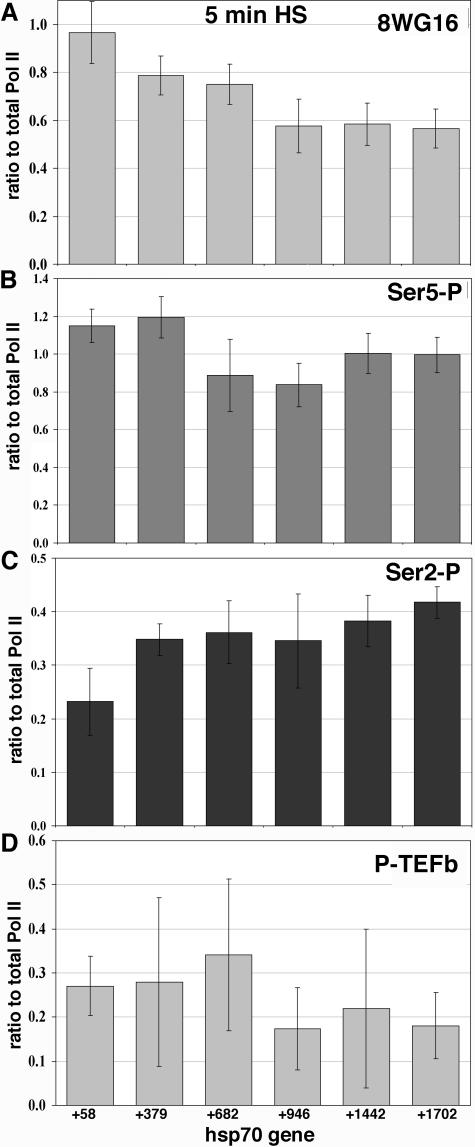
Ratio of Ser5-P to total Pol II is constant across hsp70 during active elongation, while ratio of Ser2-P to total Pol II increases.
While reliable quantitative comparisons of different Pol II forms cannot be made due to uncertainties in the efficiencies with which different antibodies precipitate their targets (even at saturating levels), the signals for a particular antibody can be compared along the gene or between different time points. Thus, by comparing total Pol II levels along hsp70 to those detected by the phosphospecific forms, we can take into account total Pol II in discussing distributions of CTD phosphorylation states. Because Pol II is consistently more predominant near the transcription start site, accounting for this will help better understand how phosphorylation changes on hsp70 during heat shock. Figure 4A compares each phosphorylation type relative to total Pol II at 5 min (which is set to a value of 1.0), when the peak level of Pol II is detected. 8WG16 signals decrease 1.7-fold relative to total Pol II as Pol II progresses through the gene (this number compares the +58 value to the +1702 value) (Fig. 4A). Importantly, Ser5-P levels, once adjusted to total Pol II levels, remain constant along the ORF (Fig. 4B). Ser2-P levels relative to that of total Pol II, on the other hand, increase slightly (1.8-fold) once Pol II progresses through the gene (Fig. 4C). This is true for all time points where Pol II has progressed through the gene. While these comparisons are useful, one must keep in mind that the ChIP signals may not scale linearly with the number of CTD epitopes because multiple epitopes exist on the Pol II CTD molecule. This may then lead to an underestimate of the quantitative differences.
FIG. 4.
Tracking the Pol II modifications and P-TEFb association with Pol II relative to total Pol II levels at the peak of factor association during heat shock. For each gene region the value for the Pol II phosphorylation state or for P-TEFb was divided by the value for total Pol II at 5 min of heat shock (thus, total Pol II/total Pol II = 1). Antibody target is indicated in the upper right-hand corner of each graph. Standard deviation for each point is shown, determined by the square root of the sum of the squares of (i) the standard deviation oftotal Pol II for that gene region and (ii) the standard deviation associated with the values of each phosphorylation state. (A) Ratio of 8WG16, unphosphorylated Pol II CTD, to total Pol II. (B) Ratio of Ser5-P signal to total Pol II. (C) Ser2-P signal ratio to total Pol II. (D) The levels of P-TEFb compared to those of total Pol II.
P-TEFb association along activated hsp70 is similar to that of Pol II.
When the 5-min time point of P-TEFb is compared to that of Pol II, the ratio of P-TEFb to Pol II detected along the gene does not change significantly (Fig. 4D). P-TEFb/Pol II ratios at the 5′ and 3′ regions of the ORF at 10 or 20 min are similar to that at 5 min, indicating that P-TEFb density does not appear to change along the gene as heat shock continues (Fig. 1G, ratios not shown).
Analysis of genes of known paused Pol II status.
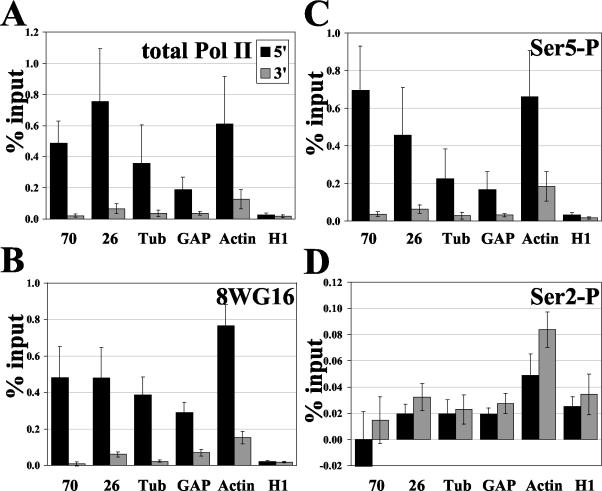
Rougvie and Lis have identified Drosophila genes other than hsp70 that harbor a paused Pol II by nuclear run-on assays (43). By using the present strategy we have analyzed several of these genes for the status of the modified forms of Pol II at the promoter in the absence of heat shock. A striking result of this analysis is that all paused Pol II candidate genes have much higher total Pol II, 8WG16, and Ser5-P levels at the 5′ end compared to those at the ORF (Fig. 5). On known pause genes hsp26, GAP, and Tub, more total Pol II is detected at the transcription start site than within the ORF (Fig. 5A). Pol II cross-linking on Tub is 9.9-fold greater at the 5′ end than the 3′ end, which agrees with previous UV cross-linking ChIP analyses (43). Actin 5C, which was difficult to assess by the run-on assay due to its alternative start sites (43), clearly has more Pol II at the promoter. Histone gene H1, which has no detectable pause in vivo, shows no significant difference in Pol II levels at the 5′ region versus those at the ORF. Results for 8WG16 and Ser5-P are similar to those of total Pol II (Fig. 5B and C). The level of Ser2-P is low, and in contrast to other modified states of Pol II, no significant differences in Ser2-P 5′ and 3′ levels are apparent on any of the genes (Fig. 5D). When total Pol II levels are taken into account, a larger fraction of Pol II is Ser2-P modified in the 3′ ORF than the promoter. This observation includes H1, although the fold difference is not the dramatic change apparent for the known pause genes.
FIG. 5.
Analysis of Pol II and its various phosphorylation states on additional genes with characterized pause states. All genes were tested in the NHS, or uninduced, state. For panels A to D, standard error n values are the same and are as follows: hsp70, n = 4; hsp26, n = 10; Tub, n = 7; GAP, n = 10; Actin5C, n = 10; and H1, n = 7. (A) ChIP analysis of total Pol II on genes in addition to hsp70. (B) Unphosphorylated Pol II CTD, 8WG16. (C) Ser5 phosphorylation. (D) Ser2 phosphorylation.
DISCUSSION
Pol II is highly regulated both at the level of recruitment to promoters and in its progress through the stages of the transcription cycle. This regulation is executed through numerous associations with other proteins as Pol II enters the promoter, melts DNA, initiates transcription, begins early elongation, and eventually matures into a productive elongation complex. Pol II undergoes additional modifications, most notably phosphorylation of the CTD of its largest subunit as it progresses from its hypophosphorylated promoter entry form to the elongation phase, where it is highly phosphorylated at residues Ser2 and Ser5. These changes in phosphorylation are proposed to influence protein association, affecting not only Pol II's elongation properties but also its association with a variety of protein complexes that process pre-mRNA. Moreover, the pattern of phosphorylation is not stagnant during the elongation phases of the cycle and may be signaling specific associations (3, 18). To define mechanisms involved in these processes in vivo, we have employed as a model the rapidly and robustly activated hsp70 gene (22).
Here we exploit technological advances of DNA-protein cross-linking and highly quantitative large-scale PCR assays to explore hsp70 activation kinetics of recruiting HSF, the critical Pol II kinase P-TEFb, and Pol II in vivo. We also examine the changes in Pol II during the first and subsequent cycles of transcription that are triggered in response to an instantaneous heat shock of Drosophila cells. HSF recruitment occurs very rapidly, detectable at the earliest assay point of 5 s of heat shock, and reaches saturation within 75 s—a result consistent with the rapid transcriptional response of heat shock genes and with previous, lower resolution assays of HSF recruitment (2, 21). The recruitment of additional hypophosphorylated Pol II to the promoter occurs with rapidity similar to that of HSF recruitment but before an increase in Pol II phosphorylation at the promoter, which occurs by 75 s. All forms of Pol II achieve a maximal level on the promoter and gene by 5 min. The progress of Pol II across the gene can be observed, and its progress fits the known rate of elongation on Drosophila hsp70, 1.2 kb/min (31). Interestingly, total Pol II levels remain greater at the transcription start site than at the ORF, even during active transcription, consistent with the observation that promoter escape remains rate-limiting even during heat shock (10). P-TEFb, a major Pol II kinase, moves across the gene at a rate similar to that of Pol II during the first burst of transcription and thereafter remains distributed over the promoter and ORF during the full 20-min time course examined. This distribution supports a model where P-TEFb contributes to Pol II phosphorylation not only at the promoter but also during most or all of the elongation phase of transcription.
The detection of Ser5 phosphorylation on a promoter-paused Pol II prior to gene induction corroborates a model where this phosphorylation is an early event involved in the transition from initiation of transcription to early phases of Pol II elongation. The mRNA associated with the paused Pol II molecule has previously been shown to be efficiently capped when long enough to allow access of the capping machinery (40). As Ser5-P has been reported to enhance Pol II association with mRNA capping machinery and capping activity (4, 5, 15, 18, 45), the Ser5-P detected on paused Pol II might help to explain the efficient capping of paused RNAs. It is important to note that earlier analyses of the hsp70 gene which determined that the paused Pol II CTD is hypophosphorylated prior to heat shock were performed with antibodies different from those used in the present study (30, 51). Importantly, the antibody generated to detect Pol IIo in those studies was directed against a peptide phosphorylated in vitro by CTK1, a yeast kinase thought to phosphorylate the CTD at serine 2 (19, 34). Thus, the previous analysis did not probe for the Ser5 phosphorylation reported here.
While P-TEFb phosphorylates the CTD primarily at Ser2, it has also been shown to recognize Ser5 as a substrate (57). Present results suggest that the Ser5 detected in the uninduced state on hsp70 is not a result of P-TEFb activity, as P-TEFb is not detected prior to gene activation (as seen in this paper and in reference 2). Ser5 is likely to be the substrate of the cdk7 component of TFIIH early in transcription. Indeed, cdk7 has been found in in vitro studies to be released earlier in the transcription cycle than P-TEFb (55, 57). In vivo, Cdk7 is required very early in the transcription cycle and contributes to the generation of the paused Pol II on the promoter-proximal region of hsp70 (45a).
Analyses of the phosphorylation status of the CTD in other organisms have found Ser5-P levels to be higher at the promoter than at the ORF (18, 48), a pattern similar to what we observe on hsp70 during active transcription. When total levels of Pol II are taken into account on hsp70, however, it appears that the level of Ser5-P remains constant along the gene. Comparatively, Komarnitsky et al. did not see a striking difference in total Pol II density along the genes analyzed (18). Soutoglou and Talianidis detected more Ser5-P at the promoter than the 3′ untranslated region of the human alpha-AT gene but also appeared to detect more total Pol II at the promoter region (48). Thus, it may be that in metazoans, or on some genes, the level of Ser5-P relative to Pol II is fairly constant along the gene. The possibility that the activity of a phosphatase may be system or gene specific is certainly plausible; for instance, heat shock of HeLa cells deactivates a CTD phosphatase (8).
Under NHS conditions, total Pol II levels were greater at the 5′ regions than at the ORFs for several genes that contain promoter-paused Pol II, while histone H1, which does not display a pause by nuclear run-on assay, shows no significant difference of 5′ and ORF Pol II signals. Greater levels of Ser5-P were also detected at the 5′ end of the genes containing paused Pol II, while levels on H1 were distributed evenly, indicating that this phosphorylation may be a general aspect of the regulatory status of a paused Pol II. This distribution of Ser5-P for the constitutively active genes Tub, GAP, and Actin5C is similar to the results of other studies which analyzed active transcription; however, Ser5-P levels on these genes are constant when standardized to total Pol II, similar to hsp70 in its active state. For these Drosophila genes, the higher level of Ser5-P at the promoter may be attributable to the presence and status of the paused Pol II, indicative of genes regulated at the level of elongation. Indeed, recent studies from Cheng et al. describe another constitutively active pause gene in human cells, dihydrofolate reductase, which shows a pattern similar to that of Ser5-P for these Drosophila genes (3).
Phosphorylation at Ser2 of the Pol II CTD may be important for processivity into active elongation and has been implicated in downstream events, including pre-mRNA splicing and 3′ mRNA processing (13, 20). Ser2-P levels were undetectable at +58 on hsp70 in the uninduced state, increased quickly at the 5′ region upon heat shock, and appeared constant through the gene in later time points (for example, 5 min). The increase in phosphorylation detected over time tracks with the recruitment of additional Pol II as well as the recruitment of P-TEFb. Taking into account total Pol II levels, there appeared to be a slight increase in Ser2-P as Pol II progressed through the ORF. This correlates with the concomitant and approximately equivalent decrease in Pol IIa. Ser2-P patterns on additional genes containing a paused Pol II, when considered relative to levels of detectable total Pol II, are significantly higher in the ORF than are those in the 5′ region. While these ratios may simply be a consequence of promoter-paused Pol II not being Ser2 phosphorylated, this result is similar to that of Komarnitsky et al., where Ser2-P was only detected in the ORF (18). These observations lead to speculation that an increase in Ser2-P may be important for cueing specific processes as Pol II proceeds through the gene. P-TEFb, the major kinase implicated in Ser2 phosphorylation, was detected concomitant with Pol II during active transcription on hsp70. While Pol II/P-TEFb ratios appear constant, a slight increase in Ser2-P occurs at the 3′ end of the gene. As the presence of the kinase is not an indicator of its activity, work presently ongoing in the laboratory on P-TEFb kinase inactivation and hsp70 gene regulation should help to better understand this process.
Lastly, analysis of immunostaining of polytene chromosomes provides independent corroboration of the higher resolution and quantitative ChIP assays and provides insight into the formation and composition of the transcription puff. Paused Pol II on hsp70 was previously detected with this method, as was Pol II along hsp70 during heat shock (51). Here we observe a modest detection of Ser5-P on the promoter prior to heat shock. During the early stages of puff formation, Pol II resolves from promoter-bound HSF. Ser2-P and Ser5-P occupy the most decondensed regions of the puff forming a halo around the heat shock loci, while HSF is more concentrated at the chromosome core at one end of the puff. Taken together, these ChIP and immunofluorescence results provide a foundation for additional temporal and spatial assignments of specific factors relative to the phosphorylation events during the activation of transcription. Perturbation of the function or activity of specific factors using genetic and drug-based approaches will provide further insight into the mechanistic role of these factors in the recruitment and modification of transcriptional machinery and in the coupling of specific transcription processes and Pol II modifications to RNA processing events.
Acknowledgments
This work was supported by National Institutes of Health grant GM25232 to J.T.L and the American Cancer Society grant PF-01-115-01-GMC to A.K.B.
We thank Carrie Davis for her helpful discussions on data processing and analysis as well as Karen Grace-Martin from the Biostatistics Department at Cornell University for statistical guidance and David Price for CycT antibody. Thanks go to W. Lee Kraus, his laboratory, and members of the Lis laboratory for scientific insight and suggestions for data presentation. The Bioresource Center at Cornell University housed the ABPrism 7900 Sequence Detection System; particular thanks go to Tom Stelick.
REFERENCES
- 1.Allison, L. A., M. Moyle, M. Shales, and C. J. Ingles. 1985. Extensive homology among the largest subunits of eukaryotic and prokaryotic RNA polymerases. Cell 42:599-610. [DOI] [PubMed] [Google Scholar]
- 2.Andrulis, E. D., E. Guzman, P. Doring, J. Werner, and J. T. Lis. 2000. High-resolution localization of Drosophila Spt5 and Spt6 at heat shock genes in vivo: roles in promoter proximal pausing and transcription elongation. Genes Dev. 14:2635-2649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cheng, C., and P. A. Sharp. 2003. RNA polymerase II accumulation in the promoter-proximal region of the dihydrofolate reductase and γ-actin genes. Mol. Cell. Biol. 23:1961-1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cho, E. J., M. S. Kobor, M. Kim, J. Greenblatt, and S. Buratowski. 2001. Opposing effects of Ctk1 kinase and Fcp1 phosphatase at Ser 2 of the RNA polymerase II C-terminal domain. Genes Dev. 15:3319-3329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cho, E. J., T. Takagi, C. R. Moore, and S. Buratowski. 1997. mRNA capping enzyme is recruited to the transcription complex by phosphorylation of the RNA polymerase II carboxy-terminal domain. Genes Dev. 11:3319-3326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Corden, J. L., D. L. Cadena, J. M. Ahearn, Jr., and M. E. Dahmus. 1985. A unique structure at the carboxyl terminus of the largest subunit of eukaryotic RNA polymerase II. Proc. Natl. Acad. Sci. USA 82:7934-7938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Costlow, N., and J. T. Lis. 1984. High-resolution mapping of DNase I-hypersensitive sites of Drosophila heat shock genes in Drosophila melanogaster and Saccharomyces cerevisiae. Mol. Cell. Biol. 4:1853-1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dubois, M. F., N. F. Marshall, V. T. Nguyen, G. K. Dahmus, F. Bonnet, M. E. Dahmus, and O. Bensaude. 1999. Heat shock of HeLa cells inactivates a nuclear protein phosphatase specific for dephosphorylation of the C-terminal domain of RNA polymerase II. Nucleic Acids Res. 27:1338-1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fong, N., and D. L. Bentley. 2001. Capping, splicing, and 3′ processing are independently stimulated by RNA polymerase II: different functions for different segments of the CTD. Genes Dev. 15:1783-1795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Giardina, C., M. Perez-Riba, and J. T. Lis. 1992. Promoter melting and TFIID complexes on Drosophila genes in vivo. Genes Dev. 6:2190-2200. [DOI] [PubMed] [Google Scholar]
- 11.Gilmour, D. S., and J. T. Lis. 1985. In vivo interactions of RNA polymerase II with genes of Drosophila melanogaster. Mol. Cell. Biol. 5:2009-2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hengartner, C. J., V. E. Myer, S. M. Liao, C. J. Wilson, S. S. Koh, and R. A. Young. 1998. Temporal regulation of RNA polymerase II by Srb10 and Kin28 cyclin-dependent kinases. Mol. Cell 2:43-53. [DOI] [PubMed] [Google Scholar]
- 13.Hirose, Y., R. Tacke, and J. L. Manley. 1999. Phosphorylated RNA polymerase II stimulates pre-mRNA splicing. Genes Dev. 13:1234-1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hirose, Y., and J. L. Manley. 1998. RNA polymerase II is an essential mRNA polyadenylation factor. Nature 395:93-96. [DOI] [PubMed] [Google Scholar]
- 15.Ho, C. K., and S. Shuman. 1999. Distinct roles for CTD Ser-2 and Ser-5 phosphorylation in the recruitment and allosteric activation of mammalian mRNA capping enzyme. Mol. Cell 3:405-411. [DOI] [PubMed] [Google Scholar]
- 16.Jiang, Y. W., P. Veschambre, H. Erdjument-Bromage, P. Tempst, J. W. Conaway, R. C. Conaway, and R. D. Kornberg. 1998. Mammalian mediator of transcriptional regulation and its possible role as an end-point of signal transduction pathways. Proc. Natl. Acad. Sci. USA 95:8538-8543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim, Y. J., S. Bjorklund, Y. Li, M. H. Sayre, and R. D. Kornberg. 1994. A multiprotein mediator of transcriptional activation and its interaction with the C-terminal repeat domain of RNA polymerase II. Cell 77:599-608. [DOI] [PubMed] [Google Scholar]
- 18.Komarnitsky, P., E. J. Cho, and S. Buratowski. 2000. Different phosphorylated forms of RNA polymerase II and associated mRNA processing factors during transcription. Genes Dev. 14:2452-2460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee, J. M., and A. L. Greenleaf. 1991. CTD kinase large subunit is encoded by CTK1, a gene required for normal growth of Saccharomyces cerevisiae. Gene Exp. 1:149-167. [PMC free article] [PubMed] [Google Scholar]
- 20.Licatalosi, D. D., G. Geiger, M. Minet, S. Schroeder, K. Cilli, J. B. McNeil, and D. L. Bentley. 2002. Functional interaction of yeast pre-mRNA 3′ end processing factors with RNA polymerase II. Mol. Cell 9:1101-1111. [DOI] [PubMed] [Google Scholar]
- 21.Lis, J. T., P. Mason, J. Peng, D. H. Price, and J. Werner. 2000. P-TEFb kinase recruitment and function at heat shock loci. Genes Dev. 14:792-803. [PMC free article] [PubMed] [Google Scholar]
- 22.Lis, J. 1998. Promoter-associated pausing in promoter architecture and postinitiation transcriptional regulation. Cold Spring Harb. Symp. Quant. Biol. 63:347-356. [DOI] [PubMed] [Google Scholar]
- 23.Lis, J. T., J. A. Simon, and C. A. Sutton. 1983. New heat shock puffs and beta-galactosidase activity resulting from transformation of Drosophila with an hsp70-lacZ hybrid gene. Cell 35:403-410. [DOI] [PubMed] [Google Scholar]
- 24.Lu, H., L. Zawel, L. Fisher, J. M. Egly, and D. Reinberg. 1992. Human general transcription factor IIH phosphorylates the C-terminal domain of RNA polymerase II. Nature 358:641-645. [DOI] [PubMed] [Google Scholar]
- 25.Marshall, N. F., J. Peng, Z. Xie, and D. H. Price. 1996. Control of RNA polymerase II elongation potential by a novel carboxyl-terminal domain kinase. J. Biol. Chem. 271:27176-27183. [DOI] [PubMed] [Google Scholar]
- 26.McCracken, S., N. Fong, K. Yankulov, S. Ballantyne, G. Pan, J. Greenblatt, S. D. Patterson, M. Wickens, and D. Bentley. 1997a. The C-terminal domain of RNA polymerase II couples mRNA processing to transcription. Nature 385:357-361. [DOI] [PubMed] [Google Scholar]
- 27.McCracken, S., N. Fong, E. Rosonina, K. Yankulov, G. Brothers, D. Siderovski, A. Hessel, S. Foster, S. Shuman, and D. L. Bentley. 1997. 5′-Capping enzymes are targeted to pre-mRNA by binding to the phosphorylated carboxy-terminal domain of RNA polymerase II. Genes Dev. 11:3306-3318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Moteki, S., and D. Price. 2002. Functional coupling of capping and transcription of mRNA. Mol. Cell 10:599-609. [DOI] [PubMed] [Google Scholar]
- 29.O'Brien, T., R. C. Wilkins, C. Giardina, and J. T. Lis. 1995. Distribution of GAGA protein on Drosophila genes in vivo. Genes Dev. 9:1098-1110. [DOI] [PubMed] [Google Scholar]
- 30.O'Brien, T., S. Hardin, A. Greenleaf, and J. T. Lis. 1994. Phosphorylation of RNA polymerase II C-terminal domain and transcriptional elongation. Nature 370:75-77. [DOI] [PubMed] [Google Scholar]
- 31.O'Brien, T., and J. T. Lis. 1993. Rapid changes in Drosophila transcription after an instantaneous heat shock. Mol. Cell. Biol. 13:3456-3463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Orphanides, G., and D. Reinberg. 2002. A unified theory of gene expression. Cell 108:439-451. [DOI] [PubMed] [Google Scholar]
- 33.Park, J. M., J. Werner, J. M. Kim, J. T. Lis, and Y. J. Kim. 2001. Mediator, not holoenzyme, is directly recruited to the heat shock promoter by HSF upon heat shock. Mol. Cell 8:9-19. [DOI] [PubMed] [Google Scholar]
- 34.Patturajan, M., N. K. Conrad, D. B. Bregman, and J. L. Corden. 1999. Yeast carboxyl-terminal domain kinase I positively and negatively regulates RNA polymerase II carboxyl-terminal domain phosphorylation. J. Biol. Chem. 274:27823-27828. [DOI] [PubMed] [Google Scholar]
- 35.Patturajan, M., R. J. Schulte, B. M. Sefton, R. Berezney, M. Vincent, O. Bensaude, S. L. Warren, and J. L. Corden. 1998. Growth-related changes in phosphorylation of yeast RNA polymerase II. J. Biol. Chem. 273:4689-4694. [DOI] [PubMed] [Google Scholar]
- 36.Price, D. H. 2000. P-TEFb, a cyclin-dependent kinase controlling elongation by RNA polymerase II. Mol. Cell. Biol. 20:2629-2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Proudfoot, N. J., A. Furger, and M. J. Dye. 2002. Integrating mRNA processing with transcription. Cell 108:501-512. [DOI] [PubMed] [Google Scholar]
- 38.Ramanathan, Y., S. M. Rajpara, S. M. Reza, E. Lees, S. Shuman, M. B. Mathews, and T. Pe'ery. 2001. Three RNA polymerase II carboxyl-terminal domain kinases display distinct substrate preferences. J. Biol. Chem. 276:10913-10920. [DOI] [PubMed] [Google Scholar]
- 39.Rasmussen, E. B., and J. T. Lis. 1995. Short transcripts of the ternary complex provide insight into RNA polymerase II elongational pausing. J. Mol. Biol. 252:522-535. [DOI] [PubMed] [Google Scholar]
- 40.Rasmussen, E. B., and J. T. Lis. 1993. In vivo transcriptional pausing and cap formation on three Drosophila heat shock genes. Proc. Natl. Acad. Sci. USA 90:7923-7927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rickert, P., W. Seghezzi, F. Shanahan, H. Cho, and E. Lees. 1996. Cyclin C/CDK8 is a novel CTD kinase associated with RNA polymerase II. Oncogene 12:2631-2640. [PubMed] [Google Scholar]
- 42.Rodriguez, C. R., E. J. Cho, M. C. Keogh, C. L. Moore, A. L. Greenleaf, and S. Buratowski. 2000. Kin28, the TFIIH-associated carboxy-terminal domain kinase, facilitates the recruitment of mRNA processing machinery to RNA polymerase II. Mol. Cell. Biol. 20:104-112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rougvie, A. E., and J. T. Lis. 1990. Postinitiation transcriptional control in Drosophila melanogaster. Mol. Cell. Biol. 10:6041-6045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rougvie, A. E., and J. T. Lis. 1988. The RNA polymerase II molecule at the 5′ end of the uninduced hsp70 gene of D. melanogaster is transcriptionally engaged. Cell 54:795-804. [DOI] [PubMed] [Google Scholar]
- 45.Schroeder, S. C., B. Schwer, S. Shuman, and D. Bentley. 2000. Dynamic association of capping enzymes with transcribing RNA polymerase II. Genes Dev. 14:2435-2440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45a.Schwartz, B. E., S. Larochelle, B. Suter, and J. T. Lis. 2003. Cdk7 is required for full activation of Drusophila heat shock genes and Pol II phosphorylation in vivo. Mol. Cell. Biol. 23:6876-6886. [DOI] [PMC free article] [PubMed]
- 46.Semeshin, V. F., E. S. Belyaeva, I. F. Zhimulev, J. T. Lis, G. Richards, and M. Bourouis. 1986. Electron microscopical analysis of Drosophila polytene chromosomes. Chromosoma 93:461-468. [Google Scholar]
- 47.Simon, J. A., C. A. Sutton, and J. T. Lis. 1985. Localization and expression of transformed DNA sequences within heat shock puffs of Drosophila melanogaster. Chromosoma 93:26-30. [DOI] [PubMed] [Google Scholar]
- 48.Soutoglou, E., and I. Talianidis. 2002. Coordination of PIC assembly and chromatin remodeling during differentiation-induced gene activation. Science 295:1901-1904. [DOI] [PubMed] [Google Scholar]
- 49.Vazquez, J., D. Pauli, and A. Tissieres. 1993. Transcriptional regulation in Drosophila during heat shock: a nuclear run-on analysis. Chromosoma 102:233-248. [DOI] [PubMed] [Google Scholar]
- 50.Wada, T., G. Orphanides, J. Hasegawa, D. K. Kim, D. Shima, Y. Yamaguchi, A. Fukuda, K. Hisatake, S. Oh, D. Reinberg, and H. Handa. 2000. FACT relieves DSIF/NELF-mediated inhibition of transcriptional elongation and reveals functional differences between P-TEFb and TFIIH. Mol. Cell 5:1067-1072. [DOI] [PubMed] [Google Scholar]
- 51.Weeks, J. R., S. E. Hardin, J. Shen, J. M. Lee, and A. L. Greenleaf. 1993. Locus-specific variation in phosphorylation state of RNA polymerase II in vivo: correlations with gene activity and transcript processing. Genes Dev. 7:2329-2344. [DOI] [PubMed] [Google Scholar]
- 52.West, M. L., and J. L. Corden. 1995. Construction and analysis of yeast RNA polymerase II CTD deletion and substitution mutations. Genetics 140:1223-1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wu, C., Y. C. Wong, and S. C. Elgin. 1979. The chromatin structure of specific genes: II. Disruption of chromatin structure during gene activity. Cell 16:807-814. [DOI] [PubMed] [Google Scholar]
- 54.Xiao, H., and J. T. Lis. 1988. Germline transformation used to define keyfeatures of heat-shock response elements. Science 239:1139-1142. [DOI] [PubMed] [Google Scholar]
- 55.Zawel, L., K. P. Kumar, and D. Reinberg. 1995. Recycling of the general transcription factors during RNA polymerase II transcription. Genes Dev. 9:1479-1490. [DOI] [PubMed] [Google Scholar]
- 56.Zhang, J., and J. L. Corden. 1991. Identification of phosphorylation sites in the repetitive carboxyl-terminal domain of the mouse RNA polymerase II largest subunit. J. Biol. Chem. 266:2290-2296. [PubMed] [Google Scholar]
- 57.Zhou, M., M. A. Halanski, M. F. Radonovich, F. Kashanchi, J. Peng, D. H. Price, and J. N. Brady. 2000. Tat modifies the activity of CDK9 to phosphorylate serine 5 of the RNA polymerase II carboxyl-terminal domain during human immunodeficiency virus type 1 transcription. Mol. Cell. Biol. 20:5077-5086. [DOI] [PMC free article] [PubMed] [Google Scholar]