Abstract
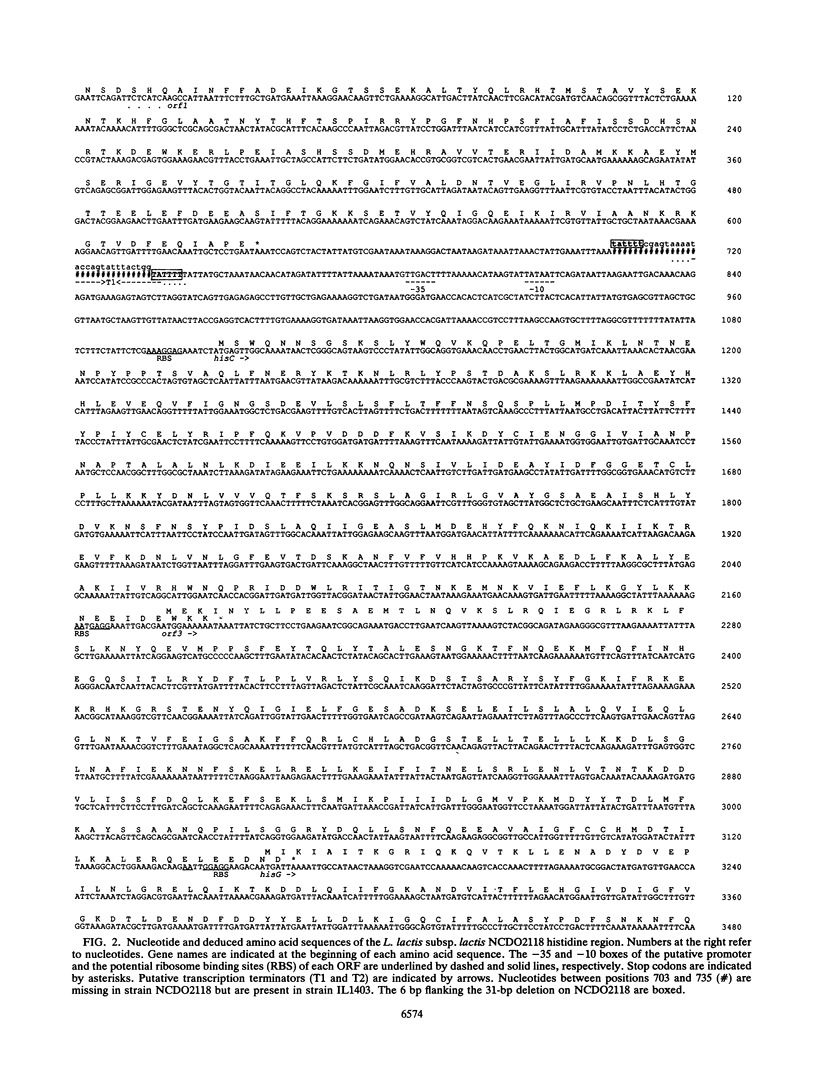
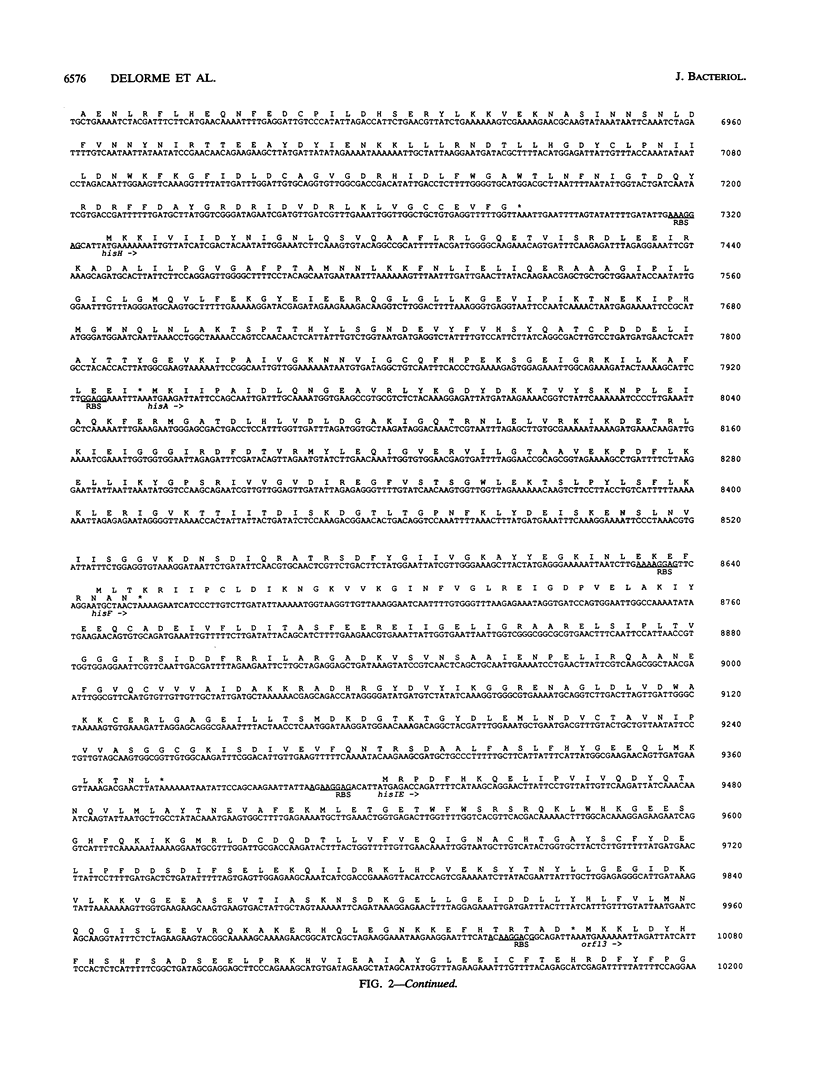
The genes of Lactococcus lactis subsp. lactis involved in histidine biosynthesis were cloned and characterized by complementation of Escherichia coli and Bacillus subtilis mutants and DNA sequencing. Complementation of E. coli hisA, hisB, hisC, hisD, hisF, hisG, and hisIE genes and the B. subtilis hisH gene (the E. coli hisC equivalent) allowed localization of the corresponding lactococcal genes. Nucleotide sequence analysis of the 11.5-kb lactococcal region revealed 14 open reading frames (ORFs), 12 of which might form an operon. The putative operon includes eight ORFs which encode proteins homologous to enzymes involved in histidine biosynthesis. The operon also contains (i) an ORF encoding a protein homologous to the histidyl-tRNA synthetases but lacking a motif implicated in synthetase activity, which suggests that it has a role different from tRNA aminoacylation, and (ii) an ORF encoding a protein that is homologous to the 3'-aminoglycoside phosphotransferases but does not confer antibiotic resistance. The remaining ORFs specify products which have no homology with proteins in the EMBL and GenBank data bases.
Full text
PDF






Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Altboum Z., Gottlieb S., Lebens G. A., Polacheck I., Segal E. Isolation of the Candida albicans histidinol dehydrogenase (HIS4) gene and characterization of a histidine auxotroph. J Bacteriol. 1990 Jul;172(7):3898–3904. doi: 10.1128/jb.172.7.3898-3904.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ames B. N., Tsang T. H., Buck M., Christman M. F. The leader mRNA of the histidine attenuator region resembles tRNAHis: possible general regulatory implications. Proc Natl Acad Sci U S A. 1983 Sep;80(17):5240–5242. doi: 10.1073/pnas.80.17.5240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bardowski J., Ehrlich S. D., Chopin A. Tryptophan biosynthesis genes in Lactococcus lactis subsp. lactis. J Bacteriol. 1992 Oct;174(20):6563–6570. doi: 10.1128/jb.174.20.6563-6570.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes W. M., Tuley E. DNA sequence changes of mutations in the histidine operon control region that decrease attenuation. J Mol Biol. 1983 Apr 15;165(3):443–459. doi: 10.1016/s0022-2836(83)80212-5. [DOI] [PubMed] [Google Scholar]
- Beckler G. S., Reeve J. N. Conservation of primary structure in the hisI gene of the archaebacterium, Methanococcus vannielii, the eubacterium Escherichia coli, and the eucaryote Saccharomyces cerevisiae. Mol Gen Genet. 1986 Jul;204(1):133–140. doi: 10.1007/BF00330200. [DOI] [PubMed] [Google Scholar]
- Bruni C. B., Carlomagno M. S., Formisano S., Paolella G. Primary and secondary structural homologies between the HIS4 gene product of Saccharomyces cerevisiae and the hisIE and hisD gene products of Escherichia coli and Salmonella typhimurium. Mol Gen Genet. 1986 Jun;203(3):389–396. doi: 10.1007/BF00422062. [DOI] [PubMed] [Google Scholar]
- Butler J. S., Springer M., Dondon J., Grunberg-Manago M. Posttranscriptional autoregulation of Escherichia coli threonyl tRNA synthetase expression in vivo. J Bacteriol. 1986 Jan;165(1):198–203. doi: 10.1128/jb.165.1.198-203.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlomagno M. S., Chiariotti L., Alifano P., Nappo A. G., Bruni C. B. Structure and function of the Salmonella typhimurium and Escherichia coli K-12 histidine operons. J Mol Biol. 1988 Oct 5;203(3):585–606. doi: 10.1016/0022-2836(88)90194-5. [DOI] [PubMed] [Google Scholar]
- Chapman L. F., Nester E. W. Gene-enzyme relationships in histidine biosynthesis in Bacillus subtilis. J Bacteriol. 1969 Mar;97(3):1444–1448. doi: 10.1128/jb.97.3.1444-1448.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiariotti L., Nappo A. G., Carlomagno M. S., Bruni C. B. Gene structure in the histidine operon of Escherichia coli. Identification and nucleotide sequence of the hisB gene. Mol Gen Genet. 1986 Jan;202(1):42–47. doi: 10.1007/BF00330514. [DOI] [PubMed] [Google Scholar]
- Chopin A., Chopin M. C., Moillo-Batt A., Langella P. Two plasmid-determined restriction and modification systems in Streptococcus lactis. Plasmid. 1984 May;11(3):260–263. doi: 10.1016/0147-619x(84)90033-7. [DOI] [PubMed] [Google Scholar]
- Conover R. K., Doolittle W. F. Characterization of a gene involved in histidine biosynthesis in Halobacterium (Haloferax) volcanii: isolation and rapid mapping by transformation of an auxotroph with cosmid DNA. J Bacteriol. 1990 Jun;172(6):3244–3249. doi: 10.1128/jb.172.6.3244-3249.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cue D., Beckler G. S., Reeve J. N., Konisky J. Structure and sequence divergence of two archaebacterial genes. Proc Natl Acad Sci U S A. 1985 Jun;82(12):4207–4211. doi: 10.1073/pnas.82.12.4207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cusack S., Härtlein M., Leberman R. Sequence, structural and evolutionary relationships between class 2 aminoacyl-tRNA synthetases. Nucleic Acids Res. 1991 Jul 11;19(13):3489–3498. doi: 10.1093/nar/19.13.3489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- EPHRATI-ELIZUR E., SRINIVASAN P. R., ZAMENHOF S. Genetic analysis, by means of transformation, of histidine linkage groups in Bacillus subtilis. Proc Natl Acad Sci U S A. 1961 Jan 15;47:56–63. doi: 10.1073/pnas.47.1.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eriani G., Delarue M., Poch O., Gangloff J., Moras D. Partition of tRNA synthetases into two classes based on mutually exclusive sets of sequence motifs. Nature. 1990 Sep 13;347(6289):203–206. doi: 10.1038/347203a0. [DOI] [PubMed] [Google Scholar]
- Fani R., Bazzicalupo M., Damiani G., Bianchi A., Schipani C., Sgaramella V., Polsinelli M. Cloning of histidine genes of Azospirillum brasilense: organization of the ABFH gene cluster and nucleotide sequence of the hisB gene. Mol Gen Genet. 1989 Apr;216(2-3):224–229. doi: 10.1007/BF00334360. [DOI] [PubMed] [Google Scholar]
- Freedman R., Gibson B., Donovan D., Biemann K., Eisenbeis S., Parker J., Schimmel P. Primary structure of histidine-tRNA synthetase and characterization of hisS transcripts. J Biol Chem. 1985 Aug 25;260(18):10063–10068. [PubMed] [Google Scholar]
- Garrick-Silversmith L., Hartman P. E. Histidine-requiring mutants of Escherichia coli K12. Genetics. 1970 Oct;66(2):231–244. doi: 10.1093/genetics/66.2.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godon J. J., Chopin M. C., Ehrlich S. D. Branched-chain amino acid biosynthesis genes in Lactococcus lactis subsp. lactis. J Bacteriol. 1992 Oct;174(20):6580–6589. doi: 10.1128/jb.174.20.6580-6589.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldschmidt E. P., Cater M. S., Matney T. S., Butler M. A., Greene A. Genetic analysis of the histidine operon in Escherichia coli K12. Genetics. 1970 Oct;66(2):219–229. doi: 10.1093/genetics/66.2.219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henner D. J., Band L., Flaggs G., Chen E. The organization and nucleotide sequence of the Bacillus subtilis hisH, tyrA and aroE genes. Gene. 1986;49(1):147–152. doi: 10.1016/0378-1119(86)90394-x. [DOI] [PubMed] [Google Scholar]
- Herbert C. J., Giles I. G., Akhtar M. The sequence of an antibiotic resistance gene from an antibiotic-producing bacterium. Homologies with transposon genes. FEBS Lett. 1983 Aug 22;160(1-2):67–71. doi: 10.1016/0014-5793(83)80937-5. [DOI] [PubMed] [Google Scholar]
- Jasin M., Regan L., Schimmel P. Modular arrangement of functional domains along the sequence of an aminoacyl tRNA synthetase. Nature. 1983 Dec 1;306(5942):441–447. doi: 10.1038/306441a0. [DOI] [PubMed] [Google Scholar]
- Johnston H. M., Barnes W. M., Chumley F. G., Bossi L., Roth J. R. Model for regulation of the histidine operon of Salmonella. Proc Natl Acad Sci U S A. 1980 Jan;77(1):508–512. doi: 10.1073/pnas.77.1.508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legerton T. L., Yanofsky C. Cloning and characterization of the multifunctional his-3 gene of Neurospora crassa. Gene. 1985;39(2-3):129–140. doi: 10.1016/0378-1119(85)90306-3. [DOI] [PubMed] [Google Scholar]
- Limauro D., Avitabile A., Cappellano C., Puglia A. M., Bruni C. B. Cloning and characterization of the histidine biosynthetic gene cluster of Streptomyces coelicolor A3(2). Gene. 1990 May 31;90(1):31–41. doi: 10.1016/0378-1119(90)90436-u. [DOI] [PubMed] [Google Scholar]
- Low B. Formation of merodiploids in matings with a class of Rec- recipient strains of Escherichia coli K12. Proc Natl Acad Sci U S A. 1968 May;60(1):160–167. doi: 10.1073/pnas.60.1.160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin P., Jullien E., Courvalin P. Nucleotide sequence of Acinetobacter baumannii aphA-6 gene: evolutionary and functional implications of sequence homologies with nucleotide-binding proteins, kinases and other aminoglycoside-modifying enzymes. Mol Microbiol. 1988 Sep;2(5):615–625. doi: 10.1111/j.1365-2958.1988.tb00070.x. [DOI] [PubMed] [Google Scholar]
- Nagai A., Ward E., Beck J., Tada S., Chang J. Y., Scheidegger A., Ryals J. Structural and functional conservation of histidinol dehydrogenase between plants and microbes. Proc Natl Acad Sci U S A. 1991 May 15;88(10):4133–4137. doi: 10.1073/pnas.88.10.4133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natsoulis G., Hilger F., Fink G. R. The HTS1 gene encodes both the cytoplasmic and mitochondrial histidine tRNA synthetases of S. cerevisiae. Cell. 1986 Jul 18;46(2):235–243. doi: 10.1016/0092-8674(86)90740-3. [DOI] [PubMed] [Google Scholar]
- Parker J., Fishman S. E. Mapping hisS, the structural gene for histidyl-transfer ribonucleic acid synthetase, in Escherichia coli. J Bacteriol. 1979 Apr;138(1):264–267. doi: 10.1128/jb.138.1.264-267.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piggot P. J., Hoch J. A. Revised genetic linkage map of Bacillus subtilis. Microbiol Rev. 1985 Jun;49(2):158–179. doi: 10.1128/mr.49.2.158-179.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putney S. D., Schimmel P. An aminoacyl tRNA synthetase binds to a specific DNA sequence and regulates its gene transcription. Nature. 1981 Jun 25;291(5817):632–635. doi: 10.1038/291632a0. [DOI] [PubMed] [Google Scholar]
- Rodriguez R. L., West R. W., Tait R. C., Jaynes J. M., Shanmugam K. T. Isolation and characterization of the hisG and hisD genes of Klebsiella pneumoniae. Gene. 1981 Dec;16(1-3):317–320. doi: 10.1016/0378-1119(81)90087-1. [DOI] [PubMed] [Google Scholar]
- Simon D., Chopin A. Construction of a vector plasmid family and its use for molecular cloning in Streptococcus lactis. Biochimie. 1988 Apr;70(4):559–566. doi: 10.1016/0300-9084(88)90093-4. [DOI] [PubMed] [Google Scholar]
- Struhl K. Nucleotide sequence and transcriptional mapping of the yeast pet56-his3-ded1 gene region. Nucleic Acids Res. 1985 Dec 9;13(23):8587–8601. doi: 10.1093/nar/13.23.8587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trieu-Cuot P., Courvalin P. Nucleotide sequence of the Streptococcus faecalis plasmid gene encoding the 3'5"-aminoglycoside phosphotransferase type III. Gene. 1983 Sep;23(3):331–341. doi: 10.1016/0378-1119(83)90022-7. [DOI] [PubMed] [Google Scholar]
- Weil C. F., Beckler G. S., Reeve J. N. Structure and organization of the hisA gene of the thermophilic archaebacterium Methanococcus thermolithotrophicus. J Bacteriol. 1987 Oct;169(10):4857–4860. doi: 10.1128/jb.169.10.4857-4860.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilbur W. J., Lipman D. J. Rapid similarity searches of nucleic acid and protein data banks. Proc Natl Acad Sci U S A. 1983 Feb;80(3):726–730. doi: 10.1073/pnas.80.3.726. [DOI] [PMC free article] [PubMed] [Google Scholar]



