Abstract
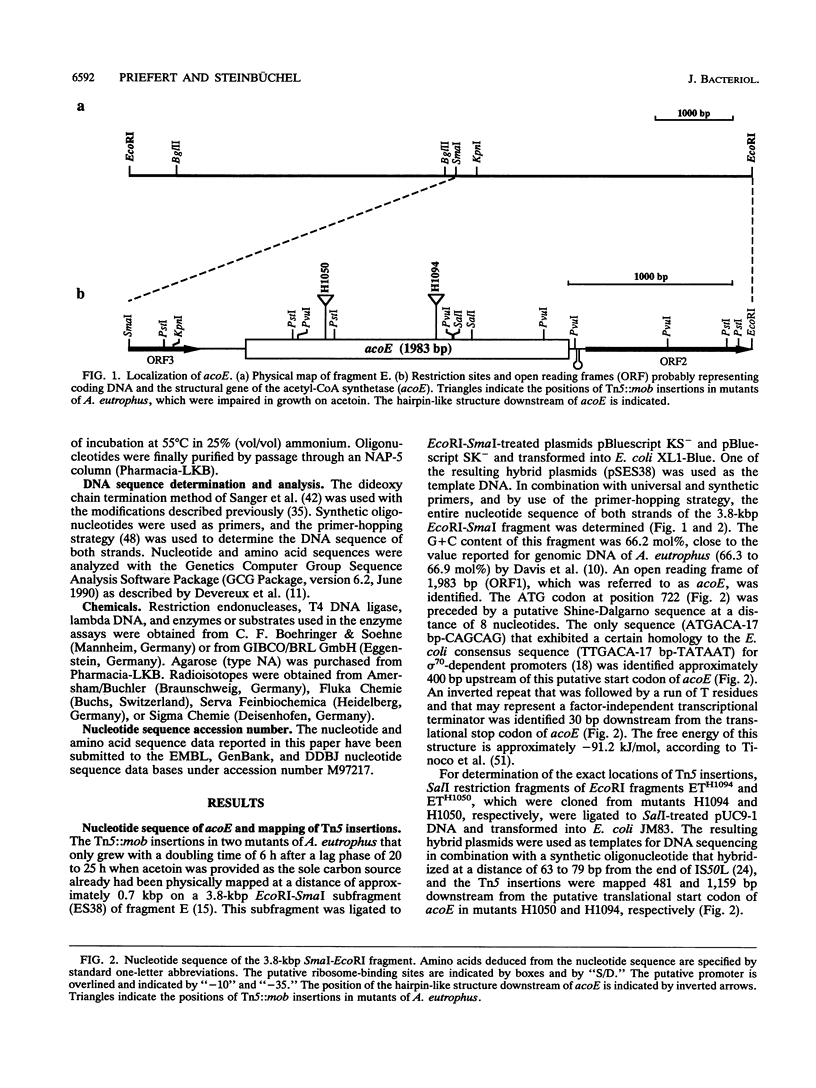
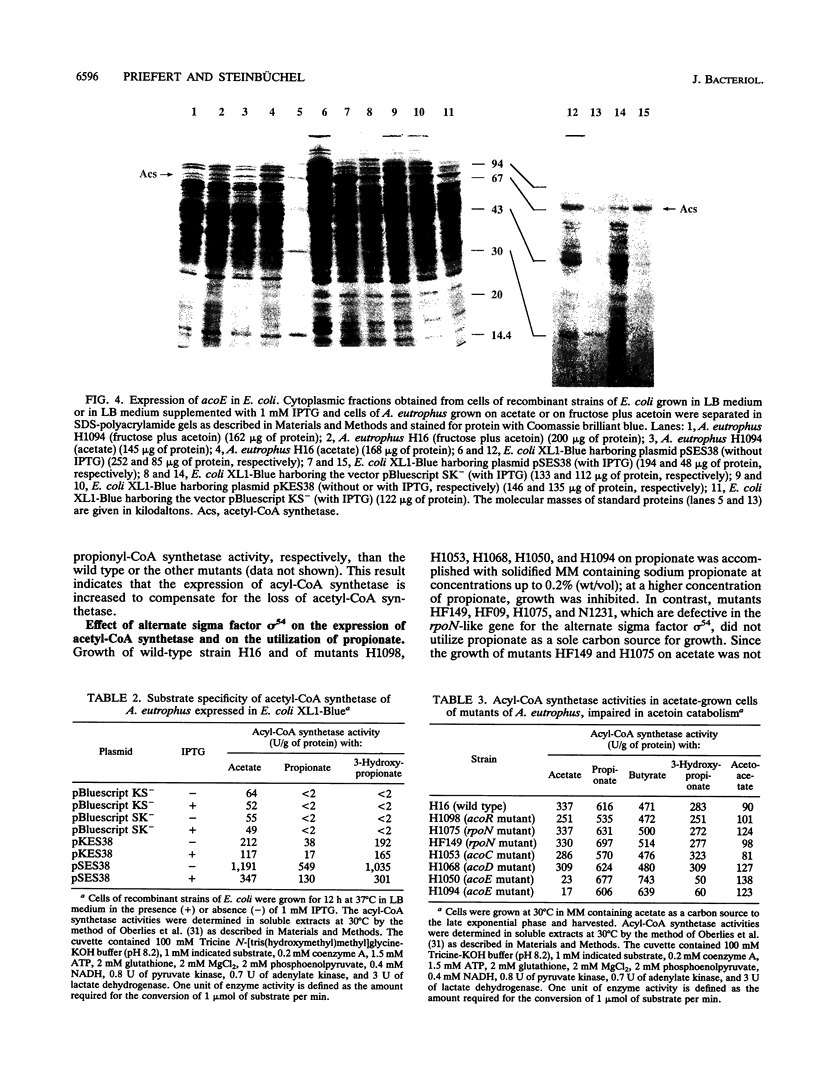
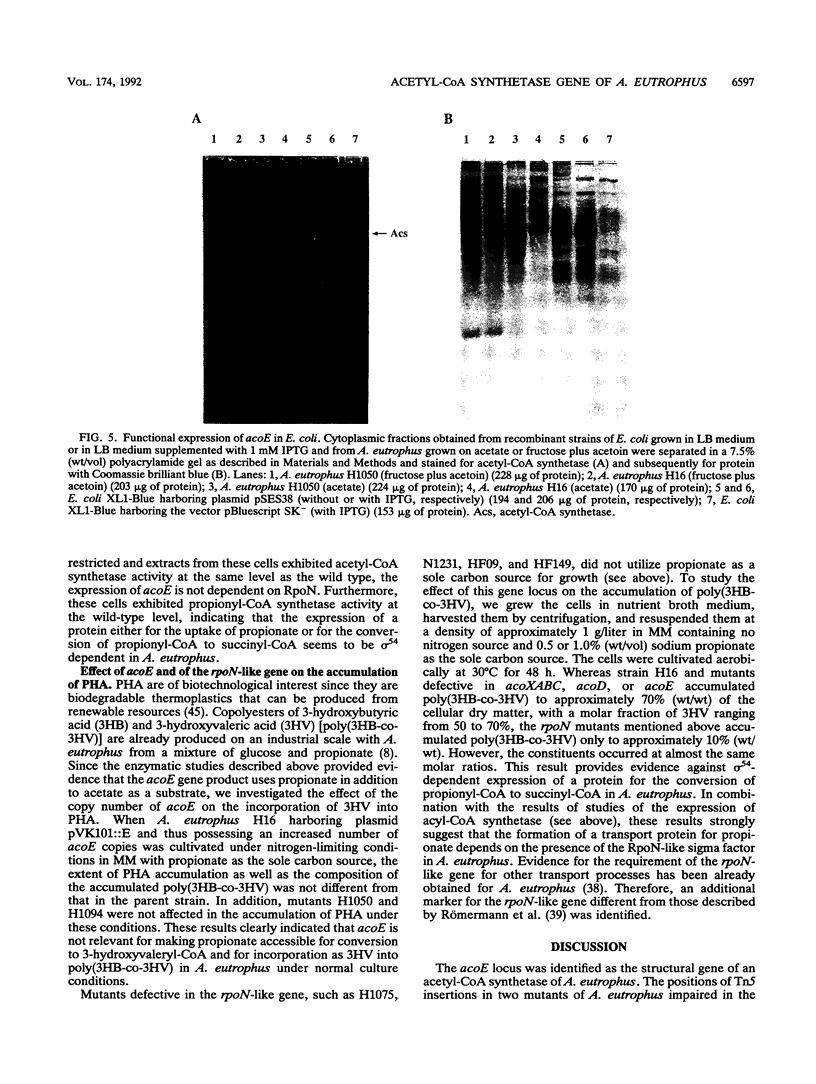
The gene locus acoE, which is involved in the utilization of acetoin in Alcaligenes eutrophus, was identified as the structural gene of an acetyl coenzyme A synthetase (acetate:coenzyme A ligase [AMP forming]; EC 6.2.1.1). This gene was localized on a 3.8-kbp SmaI-EcoRI subfragment of an 8.1-kbp EcoRI restriction fragment (fragment E) that was cloned recently (C. Fründ, H. Priefert, A. Steinbüchel, and H. G. Schlegel, J. Bacteriol. 171:6539-6548, 1989). The 1,983 bp acoE gene encoded a protein with a relative molecular weight of 72,519, and it was preceded by a putative Shine-Dalgarno sequence. A comparison analysis of the amino acid sequence deduced from acoE revealed a high degree of homology to primary structures of acetyl coenzyme A synthetases from other sources (amounting to up to 50.5% identical amino acids). Tn5 insertions in two transposon-induced mutants of A. eutrophus, that were impaired in the catabolism of acetoin were mapped 481 and 1,159 bp downstream from the translational start codon of acoE. The expression of acoE in Escherichia coli led to the formation of an acyl coenzyme A synthetase that accepted acetate as the preferred substrate (100% relative activity) but also reacted with propionate (46%) and hydroxypropionate (87%); fatty acids consisting of four or more carbon atoms were not accepted. In addition, evidence for the presence of a second acyl coenzyme A synthetase was obtained; this enzyme exhibited a different substrate specificity. The latter enzyme is obviously required for the activation of propionate, e.g., during the formation of the storage compound poly(3-hydroxybutyric acid-co-3-hydroxyvaleric acid) when propionate is provided as the sole carbon source. An analysis of mutants provided evidence that the expression of the uptake protein for propionate depends on the presence of alternate sigma factor sigma 54.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Albano M., Breitling R., Dubnau D. A. Nucleotide sequence and genetic organization of the Bacillus subtilis comG operon. J Bacteriol. 1989 Oct;171(10):5386–5404. doi: 10.1128/jb.171.10.5386-5404.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armitt S., McCullough W., Roberts C. F. Analysis of acetate non-utilizing (acu) mutants in Aspergillus nidulans. J Gen Microbiol. 1976 Feb;92(2):263–282. doi: 10.1099/00221287-92-2-263. [DOI] [PubMed] [Google Scholar]
- Bibb M. J., Findlay P. R., Johnson M. W. The relationship between base composition and codon usage in bacterial genes and its use for the simple and reliable identification of protein-coding sequences. Gene. 1984 Oct;30(1-3):157–166. doi: 10.1016/0378-1119(84)90116-1. [DOI] [PubMed] [Google Scholar]
- Brown T. D., Jones-Mortimer M. C., Kornberg H. L. The enzymic interconversion of acetate and acetyl-coenzyme A in Escherichia coli. J Gen Microbiol. 1977 Oct;102(2):327–336. doi: 10.1099/00221287-102-2-327. [DOI] [PubMed] [Google Scholar]
- Connerton I. F., Fincham J. R., Sandeman R. A., Hynes M. J. Comparison and cross-species expression of the acetyl-CoA synthetase genes of the Ascomycete fungi, Aspergillus nidulans and Neurospora crassa. Mol Microbiol. 1990 Mar;4(3):451–460. doi: 10.1111/j.1365-2958.1990.tb00611.x. [DOI] [PubMed] [Google Scholar]
- Devereux J., Haeberli P., Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 1):387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eggen R. I., Geerling A. C., Boshoven A. B., de Vos W. M. Cloning, sequence analysis, and functional expression of the acetyl coenzyme A synthetase gene from Methanothrix soehngenii in Escherichia coli. J Bacteriol. 1991 Oct;173(20):6383–6389. doi: 10.1128/jb.173.20.6383-6389.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flavell R. B., Fincham J. R. Acetate-onutilizing mutants of Neurospora crassa. I. Mutant isolation, complementation studies, and linkage relationships. J Bacteriol. 1968 Mar;95(3):1056–1062. doi: 10.1128/jb.95.3.1056-1062.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedrich B., Hogrefe C., Schlegel H. G. Naturally occurring genetic transfer of hydrogen-oxidizing ability between strains of Alcaligenes eutrophus. J Bacteriol. 1981 Jul;147(1):198–205. doi: 10.1128/jb.147.1.198-205.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fründ C., Priefert H., Steinbüchel A., Schlegel H. G. Biochemical and genetic analyses of acetoin catabolism in Alcaligenes eutrophus. J Bacteriol. 1989 Dec;171(12):6539–6548. doi: 10.1128/jb.171.12.6539-6548.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanahan D. Studies on transformation of Escherichia coli with plasmids. J Mol Biol. 1983 Jun 5;166(4):557–580. doi: 10.1016/s0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- Hanna Z., Fregeau C., Préfontaine G., Brousseau R. Construction of a family of universal expression plasmid vectors. Gene. 1984 Oct;30(1-3):247–250. doi: 10.1016/0378-1119(84)90128-8. [DOI] [PubMed] [Google Scholar]
- Hawley D. K., McClure W. R. Compilation and analysis of Escherichia coli promoter DNA sequences. Nucleic Acids Res. 1983 Apr 25;11(8):2237–2255. doi: 10.1093/nar/11.8.2237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogrefe C., Römermann D., Friedrich B. Alcaligenes eutrophus hydrogenase genes (Hox). J Bacteriol. 1984 Apr;158(1):43–48. doi: 10.1128/jb.158.1.43-48.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue T., Sunagawa M., Mori A., Imai C., Fukuda M., Takagi M., Yano K. Cloning and sequencing of the gene encoding the 72-kilodalton dehydrogenase subunit of alcohol dehydrogenase from Acetobacter aceti. J Bacteriol. 1989 Jun;171(6):3115–3122. doi: 10.1128/jb.171.6.3115-3122.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jendrossek D., Steinbüchel A., Schlegel H. G. Alcohol dehydrogenase gene from Alcaligenes eutrophus: subcloning, heterologous expression in Escherichia coli, sequencing, and location of Tn5 insertions. J Bacteriol. 1988 Nov;170(11):5248–5256. doi: 10.1128/jb.170.11.5248-5256.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jendrossek D., Steinbüchel A., Schlegel H. G. Three different proteins exhibiting NAD-dependent acetaldehyde dehydrogenase activity from Alcaligenes eutrophus. Eur J Biochem. 1987 Sep 15;167(3):541–548. doi: 10.1111/j.1432-1033.1987.tb13371.x. [DOI] [PubMed] [Google Scholar]
- Jetten M. S., Stams A. J., Zehnder A. J. Isolation and characterization of acetyl-coenzyme A synthetase from Methanothrix soehngenii. J Bacteriol. 1989 Oct;171(10):5430–5435. doi: 10.1128/jb.171.10.5430-5435.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorgensen R. A., Rothstein S. J., Reznikoff W. S. A restriction enzyme cleavage map of Tn5 and location of a region encoding neomycin resistance. Mol Gen Genet. 1979;177(1):65–72. doi: 10.1007/BF00267254. [DOI] [PubMed] [Google Scholar]
- Krüger N., Steinbüchel A. Identification of acoR, a regulatory gene for the expression of genes essential for acetoin catabolism in Alcaligenes eutrophus H16. J Bacteriol. 1992 Jul;174(13):4391–4400. doi: 10.1128/jb.174.13.4391-4400.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LOWRY O. H., ROSEBROUGH N. J., FARR A. L., RANDALL R. J. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951 Nov;193(1):265–275. [PubMed] [Google Scholar]
- Lienhard G. E., Secemski I. I. P 1 ,P 5 -Di(adenosine-5')pentaphosphate, a potent multisubstrate inhibitor of adenylate kinase. J Biol Chem. 1973 Feb 10;248(3):1121–1123. [PubMed] [Google Scholar]
- Lundie L. L., Jr, Ferry J. G. Activation of acetate by Methanosarcina thermophila. Purification and characterization of phosphotransacetylase. J Biol Chem. 1989 Nov 5;264(31):18392–18396. [PubMed] [Google Scholar]
- Nimmo H. G., Nimmo G. A. A general method for the localization of enzymes that produce phosphate, pyrophosphate, or CO2 after polyacrylamide gel electrophoresis. Anal Biochem. 1982 Mar 15;121(1):17–22. doi: 10.1016/0003-2697(82)90551-6. [DOI] [PubMed] [Google Scholar]
- Oberlies G., Fuchs G., Thauer R. K. Acetate thiokinase and the assimilation of acetate in methanobacterium thermoautotrophicum. Arch Microbiol. 1980 Dec;128(2):248–252. doi: 10.1007/BF00406167. [DOI] [PubMed] [Google Scholar]
- Oppermann F. B., Schmidt B., Steinbüchel A. Purification and characterization of acetoin:2,6-dichlorophenolindophenol oxidoreductase, dihydrolipoamide dehydrogenase, and dihydrolipoamide acetyltransferase of the Pelobacter carbinolicus acetoin dehydrogenase enzyme system. J Bacteriol. 1991 Jan;173(2):757–767. doi: 10.1128/jb.173.2.757-767.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel S. S., Walt D. R. Substrate specificity of acetyl coenzyme A synthetase. J Biol Chem. 1987 May 25;262(15):7132–7134. [PubMed] [Google Scholar]
- Peoples O. P., Sinskey A. J. Poly-beta-hydroxybutyrate biosynthesis in Alcaligenes eutrophus H16. Characterization of the genes encoding beta-ketothiolase and acetoacetyl-CoA reductase. J Biol Chem. 1989 Sep 15;264(26):15293–15297. [PubMed] [Google Scholar]
- Priefert H., Hein S., Krüger N., Zeh K., Schmidt B., Steinbüchel A. Identification and molecular characterization of the Alcaligenes eutrophus H16 aco operon genes involved in acetoin catabolism. J Bacteriol. 1991 Jul;173(13):4056–4071. doi: 10.1128/jb.173.13.4056-4071.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Priefert H., Krüger N., Jendrossek D., Schmidt B., Steinbüchel A. Identification and molecular characterization of the gene coding for acetaldehyde dehydrogenase II (acoD) of Alcaligenes eutrophus. J Bacteriol. 1992 Feb;174(3):899–907. doi: 10.1128/jb.174.3.899-907.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pries A., Priefert H., Krüger N., Steinbüchel A. Identification and characterization of two Alcaligenes eutrophus gene loci relevant to the poly(beta-hydroxybutyric acid)-leaky phenotype which exhibit homology to ptsH and ptsI of Escherichia coli. J Bacteriol. 1991 Sep;173(18):5843–5853. doi: 10.1128/jb.173.18.5843-5853.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ROSE I. A., GRUNBERG-MANAGO M., KOREY S. R., OCHOA S. Enzymatic phosphorylation of acetate. J Biol Chem. 1954 Dec;211(2):737–756. [PubMed] [Google Scholar]
- Römermann D., Warrelmann J., Bender R. A., Friedrich B. An rpoN-like gene of Alcaligenes eutrophus and Pseudomonas facilis controls expression of diverse metabolic pathways, including hydrogen oxidation. J Bacteriol. 1989 Feb;171(2):1093–1099. doi: 10.1128/jb.171.2.1093-1099.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SCHLEGEL H. G., KALTWASSER H., GOTTSCHALK G. [A submersion method for culture of hydrogen-oxidizing bacteria: growth physiological studies]. Arch Mikrobiol. 1961;38:209–222. [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strauss E. C., Kobori J. A., Siu G., Hood L. E. Specific-primer-directed DNA sequencing. Anal Biochem. 1986 Apr;154(1):353–360. doi: 10.1016/0003-2697(86)90536-1. [DOI] [PubMed] [Google Scholar]
- Thauer R. K., Möller-Zinkhan D., Spormann A. M. Biochemistry of acetate catabolism in anaerobic chemotrophic bacteria. Annu Rev Microbiol. 1989;43:43–67. doi: 10.1146/annurev.mi.43.100189.000355. [DOI] [PubMed] [Google Scholar]
- Timm A., Steinbüchel A. Formation of polyesters consisting of medium-chain-length 3-hydroxyalkanoic acids from gluconate by Pseudomonas aeruginosa and other fluorescent pseudomonads. Appl Environ Microbiol. 1990 Nov;56(11):3360–3367. doi: 10.1128/aem.56.11.3360-3367.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tinoco I., Jr, Borer P. N., Dengler B., Levin M. D., Uhlenbeck O. C., Crothers D. M., Bralla J. Improved estimation of secondary structure in ribonucleic acids. Nat New Biol. 1973 Nov 14;246(150):40–41. doi: 10.1038/newbio246040a0. [DOI] [PubMed] [Google Scholar]
- Tran-Betcke A., Warnecke U., Böcker C., Zaborosch C., Friedrich B. Cloning and nucleotide sequences of the genes for the subunits of NAD-reducing hydrogenase of Alcaligenes eutrophus H16. J Bacteriol. 1990 Jun;172(6):2920–2929. doi: 10.1128/jb.172.6.2920-2929.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trüper H. G. Tricarboxylic acid cycle and related enzymes in Hydrogenomonas strain H16G+ grown on various carbon sources. Biochim Biophys Acta. 1965 Dec 16;111(2):565–568. doi: 10.1016/0304-4165(65)90074-7. [DOI] [PubMed] [Google Scholar]
- Vieira J., Messing J. The pUC plasmids, an M13mp7-derived system for insertion mutagenesis and sequencing with synthetic universal primers. Gene. 1982 Oct;19(3):259–268. doi: 10.1016/0378-1119(82)90015-4. [DOI] [PubMed] [Google Scholar]
- WALDE E. [Studies on growth and synthesis of stored substance by Hydrogenomonas]. Arch Mikrobiol. 1962;43:109–137. [PubMed] [Google Scholar]