Abstract
The mammalian unfolded protein response (UPR) protects the cell against the stress of misfolded proteins in the endoplasmic reticulum (ER). We have investigated here the contribution of the UPR transcription factors XBP-1, ATF6α, and ATF6β to UPR target gene expression. Gene profiling of cell lines lacking these factors yielded several XBP-1-dependent UPR target genes, all of which appear to act in the ER. These included the DnaJ/Hsp40-like genes, p58IPK, ERdj4, and HEDJ, as well as EDEM, protein disulfide isomerase-P5, and ribosome-associated membrane protein 4 (RAMP4), whereas expression of BiP was only modestly dependent on XBP-1. Surprisingly, given previous reports that enforced expression of ATF6α induced a subset of UPR target genes, cells deficient in ATF6α, ATF6β, or both had minimal defects in upregulating UPR target genes by gene profiling analysis, suggesting the presence of compensatory mechanism(s) for ATF6 in the UPR. Since cells lacking both XBP-1 and ATF6α had significantly impaired induction of select UPR target genes and ERSE reporter activation, XBP-1 and ATF6α may serve partially redundant functions. No UPR target genes that required ATF6β were identified, nor, in contrast to XBP-1 and ATF6α, did the activity of the UPRE or ERSE promoters require ATF6β, suggesting a minor role for it during the UPR. Collectively, these results suggest that the IRE1/XBP-1 pathway is required for efficient protein folding, maturation, and degradation in the ER and imply the existence of subsets of UPR target genes as defined by their dependence on XBP-1. Further, our observations suggest the existence of additional, as-yet-unknown, key regulators of the UPR.
Translocation of newly synthesized peptides across the endoplasmic reticulum (ER) membrane and their subsequent folding, maturation, and transport relies on an elegant signaling system called the unfolded protein response (UPR) (28). Both surface and secreted proteins are synthesized in the ER, where they need to fold and assemble prior to being transported. Eukaryotic cells respond to the presence of unfolded proteins by upregulating the transcription of genes encoding ER resident protein chaperones such as the glucose-regulated BiP/Grp78 and Grp94 that assist in protein folding (16, 17). Since the ER and the nucleus are located in separate compartments of the cell, the unfolded protein signal must be sensed in the lumen of the ER and transferred across the ER membrane to be received by the nucleus. The UPR has been designed by the cell to do this (5). First discovered in yeast, the UPR has now been described as well in Caenorhabditis elegans and in mammalian cells (2, 5, 7, 45, 64). Upon sensing unfolded proteins, an ER transmembrane endoribonuclease and kinase called IRE1 oligomerizes and is activated by autophosphorylation and uses its endoribonuclease activity to excise an intron from the mRNA of the transcription factor Hac1p in yeast or its mammalian homologue, XBP-1 (2, 23, 38, 43, 45, 48, 51, 52, 58, 64). This splicing event results in the conversion of a 267-amino-acid XBP-1u encoded by unspliced XBP-1 mRNA to a 371-amino-acid XBP-1s by spliced XBP-1 mRNA in murine cells. XBP-1s then translocates into the nucleus, where it binds to its target sequence in the regulatory regions of the chaperone genes to induce their transcription (2, 23, 45, 64). Only the spliced form of the XBP-1s protein is an active transcription factor, while XBP-1u has no transactivation ability (14, 64).
Although yeast cells rely entirely on the IRE1/Hac1p signaling pathway, mammalian cells have evolved at least two additional strategies to ensure proper folding of proteins. The second ER transmembrane component of the mammalian UPR is a basic region/leucine zipper transcription factor called ATF6α that is constitutively expressed in an inactive form in the membrane of the ER. Activation in response to ER stress results in proteolytic cleavage of its N-terminal cytoplasmic domain by the S2P serine protease to produce a potent transcriptional activator of chaperone genes (12, 25, 44, 61, 62, 64). The recently described ATF6β is closely related structurally to ATF6α and posited to be involved in the UPR (11, 66). The third pathway acts at the level of posttranscriptional control of protein synthesis. An ER transmembrane component, PEK/PERK, related to PKR (interferon-induced double-stranded RNA-activated protein kinase) is a serine/threonine protein kinase that acts in the cytoplasm to phosphorylate eukaryotic initiation factor-2α (eIF2α). Phosphorylation of eIF2α results in translation attenuation in response to ER stress (9, 47).
In bacteria (Escherichia coli), chaperone functions are carried out by the DnaK gene (41). However, the basal ATPase activity of DnaK is too weak to be entirely responsible for assisted protein folding. Two accessory proteins, DnaJ and GrpE, greatly enhance the chaperone functions of DnaK. DnaJ activates ATP hydrolysis whereas GrpE, a nucleotide-exchange factor, increases the rate of ADP release (1, 41). Hsp70 family proteins, including BiP/Grp78, which is a representative ER localizing HSP70 member, perform the DnaK function in mammalian cells. A family of mammalian DnaJ/Hsp40-like proteins has recently been identified that are presumed to carry out the accessory functions. Two of them, Erdj4 and p58IPK, were shown to be induced by ER stress, localize to the ER, and modulate HSP70 activity (3, 32, 60). ERdj4 has recently been shown to stimulate the ATPase activity of BiP and to suppress ER stress-induced cell death (21, 46).
The regulation of mammalian UPR chaperone genes is not well understood. We used DNA microarray analysis to search for genes regulated by XBP-1 and ATF6α and -β. These data demonstrate that there are subsets of chaperone genes as defined largely by dependence on XBP-1 and provide genetic connections between XBP-1 and the two other known UPR signaling pathways, PERK and ATF6.
MATERIALS AND METHODS
Cell culture and cell lines.
293T and mouse embryo fibroblast (MEF) cells derived from wt and XBP-1−/− embryos were cultured in Dulbecco modified Eagle medium supplemented with 10% fetal calf serum (HyClone Laboratories). MEF-tet-off cells (Clontech) were maintained in the same medium with the addition of 100 μg of G418/ml and 1 μg of doxycycline/ml. XBP-1s inducible cells were obtained by transfecting MEF-tet-off cells with the TREhyg-XBP-1s plasmid, followed by selection in the presence of 400 μg of hygromycin B/ml. Several clones were tested for doxycycline-dependent XBP-1s expression, and one was selected for further experiments. MEF-dn-XBP cells were generated by transfecting MEF-tet-off cells with the TREhyg-dn-XBP plasmid. Because dn-XBP did not affect cell viability and growth, MEF-dn-XBP cells were maintained in medium without doxycycline. iATF6α and iATF6β MEF cells were generated by cotransfecting wild-type (wt) MEF cells with U6-iATF6α and cmv-puromycin or transducing cells with retroviruses containing each RNAi vector. The IRE1α-deficient MEFs were kindly provided by David Ron.
GeneChip analysis.
Total RNA was isolated from MEF cells with Trizol reagent (Invitrogen, Carlsbad, Calif.). cDNA synthesis, hybridization, and laser scanning of the array were carried out at the Gene Array Technology Center (Brigham and Women's Hospital, Boston, Mass.) with MG-U74A GeneChips that had 6,000 functionally characterized sequences and 6,000 expressed sequence tags (ESTs) from the UniGene database (Affymetrix, Santa Clara, Calif.) as recommended by the manufacturer. The data analysis was performed by using Affymetrix GeneChip 3.1 software under default parameter setting.
Northern and Western blot analysis.
Total RNA was prepared by using Trizol reagent, electrophoresed on 1.2% agarose-6% formaldehyde gels, and then transferred onto GeneScreen Plus membrane (NEN). 32P-radiolabeled probes were prepared by using the RediPrime II labeling system (Amersham-Pharmacia). Template DNAs for the probes were cut out by using appropriate restriction enzymes from the cDNA-containing plasmid (XBP-1, 15-830 of the murine coding region) or EST clones from the American Type Culture Collection (CHOP, IMAGE:5863055; ERdj4, IMAGE:1920927; p58IPK probe A, IMAGE:9001935; p58IPK probe B, IMAGE:2646147; ATF6α, IMAGE:4503659; MGP, IMAGE:4990627; EDEM, IMAGE:5324660; protein disulfide isomerase-related protein P5 [PDI-P5], IMAGE:2645183; ribosome-associated membrane protein 4 [RAMP4], IMAGE:3489738; and Armet, IMAGE:4983660). Grp94 and BiP probes were kindly provided by R. J. Kaufman, University of Michigan, and the HEDJ probe was kindly provided by L. Hendershot, St. Jude Children's Research Hospital, Memphis, Tenn. Probe hybridization was performed with Ultrahyb buffer as recommended by the manufacturer (Ambion). MEF cells were lysed in radioimmunoprecipitation assay buffer (50 mM Tris [pH 7.4], 150 mM NaCl, 1 mM EDTA, 1% Triton X-100, 1% sodium deoxycholate, 0.1% sodium dodecyl sulfate). Lysates and immunoprecipitates were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis and transferred to Hybond P membrane (Amersham-Pharmacia). Blots were revealed by using anti-XBP-1 (Santa Cruz Biotechnology), anti-ATF6β, or anti-ATF6α (K. Mori, Kyoto University, Kyoto, Japan) antibodies by standard procedures.
Plasmid construction and transient-transfection assays.
To make 4xXBPGL3, two complementary oligonucleotides containing the XBP-1 binding site 5′-CGCG(TGGATGACGTGTACA)4-3′ and 5′-GATC(TGTACACGTCATCCA)4-3′ were annealed and ligated to the −40-Luc plasmid (22) digested by MluI and BglII. The UPRE reporter was constructed by inserting annealed oligonucleotides containing two UPRE motifs (37), 5′-cgcgtcaCCAATcggaggcctCCACGaccaCCAATcggaggcctCCACGac-3′, to the −40-Luc plasmid between the MluI and XhoI sites (22). UPRE reporter (5xATF6GL3), pCGNATF6, and pCGNAF6(1-373) were previously described (57). The pCDNA3.1 (Clontech)-driven expression vectors for mouse XBP-1s and XBP-1u/s were described elsewhere (14). pCDNA-dn-XBP was constructed by removing the region downstream of the EcoRV site of XBP-1s cDNA in the pCDNA-XBP-1s plasmid. TREhyg-XBP-1s and TREhyg-dn-XBP were constructed by inserting the XBP-1s cDNA and DN-XBP, respectively, into TRE2hyg (Clontech) between the PvuII and SalI sites. A 0.5-kb fragment of the ERdj4 promoter was PCR amplified from a C57BL/6 mouse genomic DNA with the primers 5′-AGGCTTGGGCTCTAATGGCCTCTCAA-3′ and 5′-CCTCCGAACGCCGAGTAGCCT-3′ and then inserted into the pGL3-Basic (Promega) plasmid between NheI and XhoI to generate ERdj4GL3. MEF cells were transfected by using the Lipofectamine2000 reagent as recommended by the manufacturer (Invitrogen). Briefly, 1 μg of DNA and 3 μl of Lipofectamine 2000 reagent were each diluted into 100 μl of OPTI-MEM, mixed, and added to cells in 12-well plates at 60,000 cells per well. After 6 h, the cells were washed and cultured for 16 h in fresh medium with or without 1 μg of tunicamycin (Tm)/ml. For dual luciferase assays, 50 or 100 ng of reporter and 10 ng of RL/cmv (Promega) plasmids were cotransfected with various amounts of effector plasmids and pCDNA3.1, which was added to give 1 μg of DNA in total. Cells were lysed in passive lysis buffer for dual luciferase assays according to the manufacturer's protocol (Promega). 293T cells plated in a 10-cm dish were transfected with 5 μg of each expression plasmid by the standard calcium phosphate method.
Knockdown of ATF6α and ATF6β.
The pBS/U6 plasmid was kindly provided by Y. Shi (Harvard University, Boston, Mass.) (50). The two complementary oligonucleotides were annealed and inserted into pBS/U6 between blunt-ended ApaI and EcoRI sites to generate U6-iATF6α (5′-GGCAGTGTCGCCTGGTGTTGaagcttCAACACCAGGCGACACTGCCCtttttg-3′ and 5′-aattcaaaaaGGGCAGTGTCGCCTGGTGTTGaagcttCAACACCAGGCGACACTGCC-3′).Similarly, an ATF6β-specific RNAi vector was constructed by inserting two complementary oligonucleotides for 5′-GGGTGGCAGAAGTCAGTTTATG-3′ into the pBS/U6 vector. To make the SGFΔU3 shuttle retroviral vector for RNAi, a polylinker (PmlI, SalI, BamHI, and MluI) was inserted between the PmlI and BamHI sites of SFG tcLucECT3 (26). The hygromycin and puromycin resistance gene expression cassettes were removed by PCR amplification from the pMCSV series vectors (Invitrogen) and inserted between the BamHI and MluI sites of SGFΔU3 to generate SGFΔU3hyg and SGFΔU3pur, respectively. Lastly, the U6 promoter-iATF cassettes were excised from the pBS/U6-driven vectors by SmaI and BamHI digestion and then inserted into SGFΔU3hyg or SGFΔU3pur between the PmlI and BamHI sites to generate the retroviral vectors for iATF6α or iATF6β with various drug selection markers. Retroviral supernatant was prepared and used to transduce MEF cells as described previously (14). Uninfected cells were removed by culturing cells in the presence of 200 μg of hygromycin B or 2 μg of puromycin/ml for more than 1 week.
RESULTS
Identification of known and novel UPR genes by DNA microarray analysis.
A genome-wide analysis in yeast revealed that a subset of genes, including ER resident chaperones and those involved in phospholipid biosynthesis and protein degradation pathways, were induced through the UPR (53). Similarly, mammalian cells also induce a variety of genes upon ER stress (10, 33, 40). To identify UPR target genes that were differentially regulated by XBP-1, ATF6α, and ATF6β, we used oligonucleotide-based gene array analysis. As a standard, RNAs from MEF cells untreated or treated with Tm for 6 h were analyzed for UPR target gene expression. Expression of ∼2.8% of the total pool of 12,000 genes analyzed was increased upon Tm treatment. The gene chips that were used in the present study consisted of oligonucleotide probe sets that are 25 bases long. Each probe set that was designed to recognize one transcript consists of 16 different oligonucleotide probe pairs. To avoid the detection of false-positive genes, Tm-inducible genes were sorted by ratio of oligonucleotide probe pairs whose values were increased upon Tm treatment, and the genes whose values were >0.8 are shown in Table 1. As expected, well-known UPR target genes, including CHOP, GADD45, Herp, BiP, and XBP-1, were significantly induced by Tm treatment, with fold inductions ranging from 4 to 27. We identified, in addition, a panel of novel UPR target genes whose roles in ER stressed cells have yet to be investigated. Interestingly, Tm treatment induced the expression of several transcription factors, including CHOP, LRG-21, XBP-1, NF-IL-3/E4BP4, and ATF4, which have leucine zipper motifs. Given the known ability of transcription factors containing the leucine zipper motif to homo- and heterodimerize, it will be of interest to test for possible interactions among these proteins.
TABLE 1.
Induction of UPR target genes in wt, XBP-1−/−, and iATF6α MEF cells by Tm treatment and in MEF-tet-off-XBP-1s cells by doxycycline removal
| Fold change in expression with: | GenBank no. | Description | |||
|---|---|---|---|---|---|
| wt | XBP-1−/− | iATF6α | XBP-1s | ||
| 27.1 | 16.7 | 8.5 | NC | X67083 | CHOP |
| 21.4 | 7.1 | 27.2 | NC | U00937 | GADD45 |
| 11 | 7.2 | 4.4 | 3.2 | AI846938 | Herp |
| 4.6 | NC | 2.2 | 1.9 | AI845538 | MGP |
| 8.4 | 7 | 5.5 | NC | AW045664 | RIKEN 2810026P18, GADD7 |
| 4.5 | 2.8 | 2.8 | 1.7 | AJ002387 | BiP |
| 4.3 | 3.3 | 2.2 | NC | AA684508 | Rnu22, RNA, U22 small nucleolar |
| 2.9 | 3.3 | 2.6 | NC | X14309 | 4F2 antigen |
| 2.8 | NC | 1.6 | NC | AA260005 | Par-4 |
| 9.8 | 5.8 | 9.1 | NC | U19118 | LRG-21 |
| 4.8 | 2.2 | 4 | NC | AI840585 | RIKEN 3110043O21 |
| 4.5 | 3.2 | 3.7 | NC | L00039 | c-myc |
| 3.6 | 3.1 | 2.6 | 2.7 | AW122364 | Armet |
| 2.9 | 3.4 | 3 | NC | AB017189 | 4F2/CD98 |
| 2.8 | 3.2 | 2.6 | NC | AI849615 | Gas5, growth arrest specific 5 |
| 2.9 | 1.8 | 1.6 | 3.1 | U28423 | p581PK |
| 8.9 | 2.4 | 7.9 | 5 | AW120711 | ERdj4 |
| 8.8 | ∼15.0 | 4.9 | 2 | AW124049 | EST, Genethonin |
| 5.2 | 3.7 | 1.9 | NC | AI852641 | Nupr1, nuclear protein 1 |
| 4 | NC | 2.9 | 2.2 | AW123880 | XBP-1 |
| 3.9 | 3.2 | 3.7 | NC | AI854851 | RIKEN 2700007P21 |
| 3.8 | 3 | 5.5 | NC | U52073 | TDD5, androgen target gene |
| 3.4 | 5.2 | 3.8 | NC | U40930 | Sqstm1, squestosome |
| 3.3 | NC | 1.8 | 2.7 | AI604013 | p581PK |
| 3.3 | 3.1 | 2.9 | NC | U83148 | NFIL3/E4BP4 |
| 2.9 | 2.3 | 2.4 | 1.5 | V00756 | Beta interferon |
| 2.8 | 2.6 | 2.6 | NC | U13371 | Kdt1, kidney cell line derived transcript I |
| 2.7 | 2.6 | NC | NC | U79550 | Slug, chicken homolog |
| 2.5 | 2.6 | 1.8 | NC | M94087 | ATF4 |
| 2.4 | NC | NC | NC | AV138783 | Gadd45b |
| 9.6 | NC | 9.2 | 5.7 | AI835630 | ERdj4 |
| 4.6 | 3.8 | 6.1 | NC | U60593 | Ndr1 |
| 4.2 | NC | 3.7 | NC | AA798624 | Ero11 |
| 4.2 | 5.3 | 6.1 | NC | M31418 | Interferon activated gene 202 |
| 4.1 | 3.6 | 4.7 | NC | AI839690 | RIKEN 1500005G05 |
| 3.7 | NC | 2.7 | NC | M95200 | Vascular endothelial growth factor |
| 2.7 | 3.1 | 2 | NC | J04627 | Methenyltetrahydrofolate cyclohydrolase |
| 2.7 | NC | 2.2 | NC | AW047899 | Nfkb2 |
| 2.4 | 2.2 | 1.9 | NC | X78709 | NRF1 |
| 2.2 | 2.1 | 2.3 | NC | AI849620 | EST |
| 2 | NC | NC | NC | J03297 | grp94 |
Values represent fold changes of expression level by Tm treatment for 6 h in wt, XBP-1−/−1, and iATF6α MEF cells by Tm or doxycycline removal in MEF-tet-off-XBP-1s cells. NC, no change. Genes were sorted by ratio of oligonucleotide probe pairs on the chip whose values were increased upon Tm treatment in wt cells, and the genes whose values were >0.8 are shown. The two XBP-1-dependent genes which were confirmed by Northern blot analysis are highlighted in boldface.
Induction of the ERdj4 and p58IPK DnaJ/Hsp40-like accessory genes upon ER stress requires XBP-1.
To investigate the requirement of XBP-1 in UPR target gene expression, MEF cells were generated from XBP-1-deficient mice (Fig. 1) (34). Treatment of wt MEFs with the proteasome inhibitor MG-132 or with Tm induced both XBP-1u and XBP-1s proteins (Fig. 1C) through protein stabilization and mRNA splicing, respectively, as demonstrated previously (64; A.-H. Lee, unpublished observations). In contrast, as expected, neither XBP-1 protein species was produced in XBP-1−/− MEF cells because of multiple stop codons derived from the neo cassette (Fig. 1A and C). We then searched for XBP-1-dependent UPR target gene expression by using gene array analysis with the RNA from the XBP-1-deficient MEF cells either untreated or treated with Tm. It has been previously shown that the expression of neither BiP nor CHOP, the prototypical UPR target genes, was affected by loss of IRE1α, raising the possibility that these genes were regulated by other UPR pathways (23, 55). Not surprisingly, therefore, most of the prototypical UPR target genes identified in wt MEF cells were normally induced in XBP-1−/− MEF cells, invoking the presence of additional UPR signaling pathways that are not dependent on XBP-1 (Table 1).
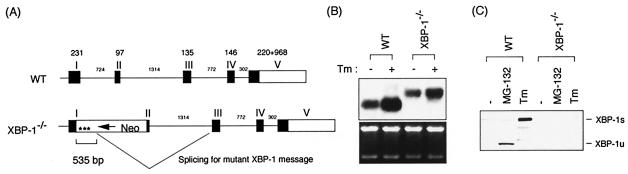
FIG. 1.
Structure of the XBP-1 gene and protein in wt and mutant cells. (A) XBP-1 locus in wt and XBP-1−/− MEF cells. Splicing of the mutant XBP-1 mRNA in XBP-1−/− cells is shown. Asterisks represent termination codons. (B) wt and XBP-1−/− MEF cells were either not treated or treated with 10 μg of Tm/ml for 6 h. XBP-1 mRNA was revealed by Northern blot analysis. (C) wt and XBP-1−/− MEF cells were treated with 10 μM MG-132 or 10 μg of Tm/ml for 6 h. XBP-1u and XBP-1s proteins were detected by Western blot analysis with anti-XBP-1 antibody.
In contrast, several known and novel UPR target genes were identified which failed to be induced by Tm in the XBP-1-null MEFs. To minimize gene array artifacts, genes whose fold induction was decreased in XBP-1−/− MEF cells, but whose expression levels were comparable between Tm-treated wt and XBP-1−/− MEF cells were excluded for further analysis. Two UPR target genes, ERdj4 and p58IPK, were not induced at all in XBP-1−/− MEF cells by this analysis, a finding that was verified by Northern blot analysis (Fig. 2A). Other genes that appeared to be noninducible in XBP-1−/− cells in this gene array analysis were false positives, as assessed by Northern blot analysis (data not shown). In time course experiments, BiP and CHOP expression were only modestly, if at all, impaired in the absence of XBP-1 (Fig. 2A). Gene array analysis suggested that MGP mRNA, an inhibitor of calcification of arteries and cartilage (27), was Tm inducible in wt MEF cells but not in XBP-1−/− MEF cells. Interestingly, however, Northern blot analysis revealed that MGP mRNA was dramatically downregulated upon ER stress, indicating that the microarray data was incorrect. (Fig. 2A). In contrast, ERdj4 and p58IPK expression was almost completely abolished in XBP-1−/− cells (Fig. 2A). There are three isoforms of p58IPK with transcripts of ∼6.5, 3.3, and 1.7 kb (20), all of which were XBP-1 dependent as assessed by using probes B and A specific for the 5′ (Fig. 2A) and 3′ regions of the gene, respectively (Fig. 2B). EST clone AI604013 represented the 3′ end of the 6.5-kb species of p58IPK mRNA, as confirmed by EST “walking” analysis. Although probe A recognized only the 6.5-kb species, probe B hybridized to all three p58IPK mRNAs, indicating that the ∼6.5-kb species shares 5′ ends with the other mRNA species.
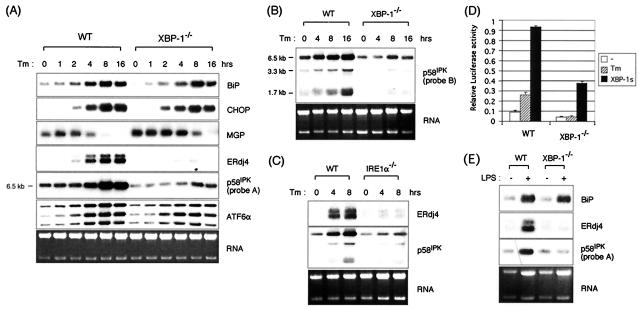
FIG. 2.
Dependence of UPR target gene expression on XBP-1. (A) wt and XBP-1−/− MEF cells were treated with 10 μg of Tm/ml for the indicated time periods. Total RNAs were isolated and subjected to Northern blot analysis. The same blot was hybridized sequentially with BiP, CHOP, MGP, ERdj4, p58IPK, ATF6α, and MGP probes. The p58IPK probe is probe A, from the 3′ end of the gene. Ethidium bromide staining of the gel before blotting is shown at the bottom for loading control. (B) All isoforms of p58IPK are XBP-1 dependent. Here, Northern blot analysis was performed with probe B, which recognizes sequences at the 5′ end of the gene. (C) XBP-1-dependent genes are also IRE1α dependent. Northern blot analysis of RNA prepared from IRE1α−/− MEFs treated with Tm for various time periods and assessed for expression of ERdj4 and p58IPK. (D) The ERdj4GL3 reporter was transfected with or without XBP-1s plasmid into wt and XBP-1−/− MEF cells. Cells were treated with Tm at 1 μg/ml for 16 h before harvesting as indicated. Luciferase activity was normalized to the Renilla activity. (E) Induction of BiP, ERdj4, and p58IPK in primary B cells by LPS. B220+ primary B cells were isolated from spleens of wt or XBP-1−/− RAG2−/− lymphoid chimeras. Cells were untreated or stimulated for 3 days with 20 μg of LPS/ml. Expression of BiP, ERdj4, and p58IPK was determined by Northern blot analysis.
The placement of the ERdj4 and p58IPK genes downstream of XBP-1 was further established by examining their expression in MEFs lacking IRE1α (Fig. 2C). Consistent with the profound effect of XBP-1 on regulating ERdj4 expression was our finding that a construct containing 0.5 kb of ERdj4 promoter sequence fused to a luciferase reporter was induced by Tm as well as by cotransfected XBP-1s (Fig. 2D). Further, the ERdj4 promoter was not induced by Tm in XBP-1−/− cells, whereas it was transactivated by cotransfected XBP-1s. Dependence of ERdj4 and p58IPK expression on XBP-1 was also tested in primary B cells in which both XBP-1 transcription and posttranscriptional splicing to the XBP-1s form are induced during terminal B-cell differentiation to plasma cells. This is functionally critical since lymphoid chimeras lacking XBP-1 fail to generate the plasma cell compartment. Both ERdj4 and p58IPK were induced in lipopolysaccharide (LPS)-stimulated wt cells but not in XBP-1-deficient B cells (Fig. 2E). In contrast, BiP was induced in both wt and XBP-1-deficient B cells although, again, there was a modest impairment in its expression in the absence of XBP-1.
We were also able to look at XBP transcripts in the mutant MEFs since the disrupted XBP-1 gene produces a transcript, 0.4 kb longer than the wt, composed of the neomycin gene and XBP-1 sequences arising from an alterative splicing event between a cryptic splice donor site in the neo cassette, inserted between exons 1 and 2, and the splice acceptor site for exon 3 (Fig. 1A and B). XBP-1 mRNAs were induced by Tm treatment in both wt and XBP-1-deficient MEF cells (Fig. 1B), similar to what was reported for IRE1α-deficient cells (23). ATF6α was also modestly induced by Tm in these murine MEFs (Fig. 2A), in contrast to what was observed in human HeLa cells (62). However, the fold induction of both XBP-1 and ATF6α was modestly decreased in the absence of XBP-1 (Fig. 1B and 2A). Although it is possible that the mutant transcript is aberrantly regulated, these results indicate some degree of autoregulation of XBP-1 as well as its cross-regulation of ATF6α.
These experiments identified both known and potentially novel UPR target genes and suggested that XBP-1 is essential for the expression of only some of them, which include the DnaJ-like accessory proteins, ERdj4 and p58IPK. It has a modest effect in regulating the expression of other known UPR target genes, such as BiP and ATF6α, and itself, and no effect on other UPR genes (CHOP and MGP).
Identification of genes induced by XBP-1s.
The minimally altered expression of some UPR target genes in XBP-1-deficient MEF cells indicates either that XBP-1 is not significantly involved in their expression or that there may be other transcription factor(s) that fully compensate for XBP-1. To examine whether XBP-1 itself is sufficient to induce these UPR target genes, we used the tet-off system to establish a cell line in which the spliced form of XBP-1, XBP-1s, is placed downstream of a tetracycline-dependent promoter. XBP-1s was induced by removing doxycycline in the culture medium for 3 days, which was the time point for maximal expression. The expression level of the exogenous XBP-1s was comparable to endogenous XBP-1s in the parental MEF cells treated with Tm (Fig. 3A). To identify genes that were induced by XBP-1s, total RNAs were prepared from MEF-tet-off-XBP-1s cells cultured in the presence or absence of doxycycline, and gene array analysis was performed (Table 1). Consistent with the results from XBP-1−/− cells, XBP-1s alone was sufficient to induce ERdj4 and p58IPK expression. Several additional UPR target genes—Herp, BiP, Aremt, AW124049 EST, and beta interferon—were identified that were induced by XBP-1s, although no change in the expression of these genes had been detected by the microarray analysis in Tm-treated XBP-1−/− cells. In contrast, several UPR target genes, including CHOP, were not significantly induced by XBP-1s, suggesting that XBP-1s is either not involved in or not sufficient for their induction. The expression of BiP, CHOP, ERdj4, and p58IPK was confirmed by Northern blot analysis (Fig. 3A). Induction of ERdj4 and p58IPK by XBP-1s overexpression was comparable to that achieved by Tm treatment, whereas BiP was only marginally induced by XBP-1s. ATF6α was also modestly induced by XBP-1s, as shown by Northern blot analysis, placing ATF6 downstream of XBP-1.
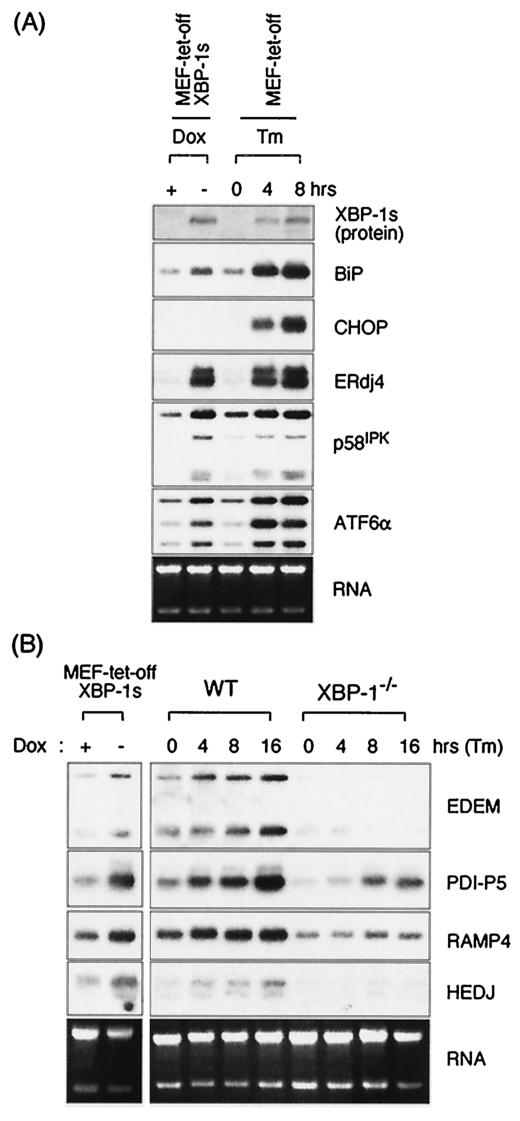
FIG. 3.
Induction of UPR target genes by XBP-1s. MEF-tet-off and MEF-tet-off-XBP-1s cells were cultured in medium containing 1 μg of doxycycline/ml. XBP-1s expression was induced by culturing the cells for 3 days in doxycycline-free medium or by treatment with Tm for the indicated times. XBP-1s protein was revealed by anti-XBP-1 antibody in Western blot analyses. Total RNA was also prepared to measure the expression level of BiP, CHOP, ERdj4, p58IPK, and ATF6α mRNA. (B) Additional XBP-1-dependent target genes identified in gene profiling experiments (Table 2) were confirmed by Northern analysis. Ethidium bromide staining of the gels before blotting is shown at the bottom.
Further, we identified several additional XBP-1 target genes—EDEM, PDI-P5, RAMP4, and HEDJ—by sorting based on the criteria of inducibility by XBP-1s (Table 2). Strikingly, their expression was induced by Tm in wt MEFs but not in XBP-1−/− MEFs, confirming their XBP-1 dependency. XBP-1-dependent expression of EDEM is consistent with the recent finding that its induction was absent in MEFs lacking IRE1α (63) (Fig. 3B). Other genes that were also identified in this microarray analysis to be inducible by XBP-1s and dependent on XBP-1 for Tm-induced expression have yet to be confirmed by Northern blotting. Collectively, these results suggest that XBP-1 is essential for the regulation of several UPR target genes, (ERdj4 p58IPK, EDEM, PDI-P5, RAMP4, and HEDJ), is modestly involved in the regulation of some UPR genes (BiP, XBP-1, and ATF6α), and is not at all required for the expression of another subset of UPR target genes. Consistent with an important function for XBP-1 in regulating UPR target genes is the failure of Tm to induce the activity of either the UPRE or ERSE luciferase reporters in the XBP-1−/− MEFs (see below, Fig. 4B).
TABLE 2.
UPR target genes induced by XBP-1s
| Fold change in expression with: | GenBank no. | Description | ||
|---|---|---|---|---|
| XBP-1s | WT | XBP-1−/− | ||
| 5.7 | 9.6 | NC | AI835630 | ERdj4 |
| 2.9 | ∼4.0 | NC | AW212878 | EDEM |
| 2.7 | 3.6 | 3.1 | AW122364 | Armet |
| 2.7 | 3.3 | NC | AI604013 | p58IPK |
| 3.2 | 2.7 | NC | U28423 | p58IPK |
| 2.2 | 1.7 | NC | AW045202 | PDI-P5 |
| 2.1 | 1.5 | NC | AI843466 | Ramp4 |
| 1.9 | 2.1 | NC | AW122551 | HEDJ |
| 2.2 | 4 | NC | AW123880 | XBP-1 |
| 1.8 | 1.7 | NC | AI117848 | mgat2 |
| 3.2 | 11 | 7.2 | AI846938 | Herp |
| 2.4 | 7.4 | 2 | D16333 | Coproporphyrinogen oxidase |
| 2 | 8.8 | ∼15.0 | AW124049 | EST |
| 1.9 | 4.6 | NC | AI845538 | MGP |
| 1.5 | 2.4 | NC | AI843342 | Splicing factor, arginine/serine-rich 1 |
| 1.9 | 1.7 | NC | AV318100 | Similar to glucosidase 1 |
| 1.7 | 1.9 | NC | M22998 | Solute carrier family 2, member 1 |
| 1.6 | 1.6 | NC | M73329 | Phospholipase Cα |
| 1.6 | 1.6 | NC | AI839280 | RIKEN cDNA 1810045K07 gene |
| 1.5 | 2.5 | NC | AW123026 | Glucosamine-phosphate N-acetyltransferase 1 |
| 2.1 | 1.5 | 2.2 | X83601 | Pentaxin related gene |
| 1.7 | 4.5 | 2.8 | AJ002387 | BiP |
| 1.6 | 1.9 | NC | AA170696 | Intercellular adhesion molecule |
| 1.6 | 2.2 | NC | AI316859 | Hypothetical protein LOC223770, BR-140L |
| 1.5 | 2.9 | 2.3 | V00756 | Interferon-related developmental regulator 1 |
| 1.5 | 1.8 | NC | X92665 | Ubiquitin-conjugating enzyme E2E 1 |
Genes that were induced by doxycycline removal in MEF-tet-off-XBP-1s cells, as well as by Tm treatment in wt MEF cells are shown. Genes were sorted by ratio of positive oligonucleotide probe pairs upon doxycycline removal in MEF-tet-off-XBP-1s cells, and the genes that were as also inducible by Tm in wt cells are shown. Values represent the fold changes in expression level. NC, no change.
FIG. 4.
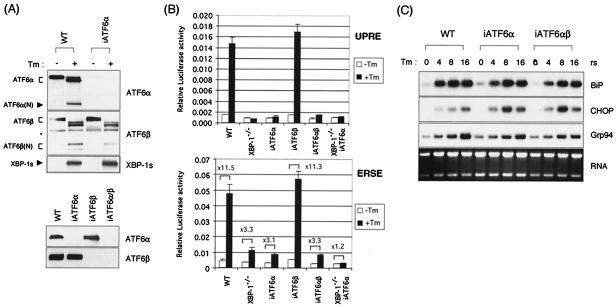
UPR target gene expression is largely unaffected in the absence of ATF6α and ATF6β. (A) Western blot analysis of iATF6α-, iATF6β-, and double iATF6α/β-expressing MEFs. MEF-iATF6α cells were generated by transfecting MEF cells with U6-iATF6α plasmid, which expresses siRNA for ATF6α under the control of the U6 promoter. MEF-iATF6β and MEF-iATF6α/β cells were generated by transducing wt and MEF-iATF6α cells with retroviruses that express iATF6β. Lysates from wt and MEF-iATF6α, MEF-iATF6β, and MEF-iATF6α/β MEFs either not treated or treated with 10 μg of Tm/ml for 6 h were analyzed for the expression of XBP-1, ATF6α, and ATF6β. An asterisk indicates a nonspecific band recognized by anti-ATF6α antibody. (B) 5xATF6GL3 (UPRE) or ERSE reporters were transfected into wt, XBP-1−/−, iATF6α, iATF6β, double iATF6α/β, and double XBP-1−/−/iATF6α MEF cells. Cells were treated with Tm at 1 μg/ml for 16 h before harvesting as indicated. Luciferase activity was normalized to Renilla activity. The fold induction of relative luciferase activity by Tm treatment compared to untreated samples is also shown. (C) Total RNA was prepared to measure the expression level of BiP, CHOP, ERdj4, p58IPK, and Grp94 mRNAs. Ethidium bromide staining of the gel before blotting is shown at the bottom.
UPR gene expression in ATF6α and ATF6β single and double deficient cells.
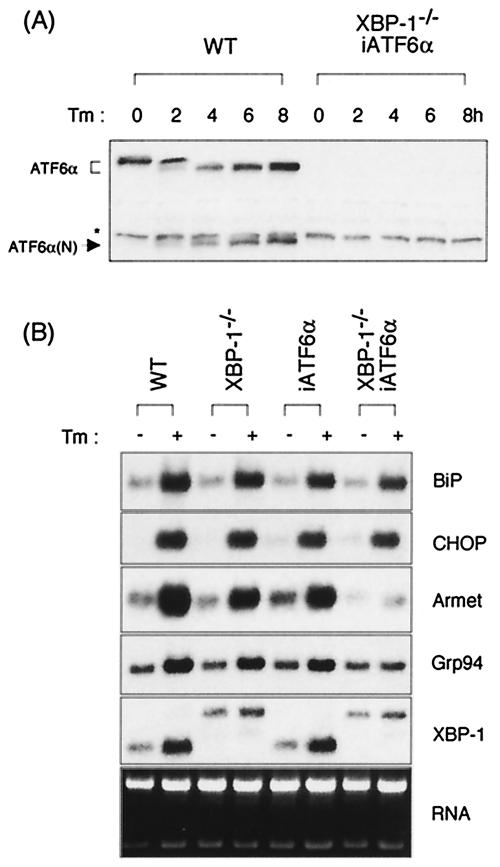
The ER transmembrane transcription factor ATF6α is proteolytically processed to release its active N-terminal region for nuclear transport upon ER stress and has been reported to autonomously induce a subset of UPR target genes, including BiP and CHOP (33, 62). Mice carrying a targeted deletion of the ATF6α gene are not available. To more directly assess the requirement of ATF6α in the UPR, we therefore “knocked-down” its expression in MEF cells by using an RNA polymerase III-driven small interfering RNA (siRNA) expression plasmid. Cotransfection of the ATF6α-specific siRNA vector with a multimerized ATF6 target site-luciferase reporter (5xATF6GL3) resulted in suppression of both ATF6-driven and Tm-evoked luciferase expression, suggesting that ATF6α mRNA had been appropriately targeted by the siRNA vector (not shown). MEF cells were therefore stably transfected with the ATF6α siRNA vector to generate cell lines in which ATF6α expression was reduced (iATF6α). Suppression of ATF6α expression was confirmed by both Northern (not shown) and Western blot analysis (Fig. 4A). In the parental wt MEF line, ATF6α protein was synthesized as an ER resident 90-kDa precursor form and cleaved by S2P proteases to generate the 50-kDa active form upon ER stress, as expected (Fig. 4A). In contrast, although a small amount of the full-length inactive protein was detected after prolonged exposure of the film (data not shown), the processed ATF6α protein was not detected in the iATF6α cells, in either the absence or the presence of Tm treatment (Fig. 4A and B). Levels of XBP-1s and ATF6β transcripts (results not shown) and protein (Fig. 4A and B) were not altered in the iATF6α MEFs. This latter point is of interest since it suggests that XBP-1 is not downstream of ATF6α as previously suggested (64, 65). Transient-transfection assays revealed that induction of the UPRE (5xATF6GL3) and ERSE reporters by Tm treatment was absent or significantly diminished in the iATF6α MEFs (Fig. 4B). We concluded that this cell line behaved in a manner consistent with a functional absence of ATF6α.
Gene array analysis was then performed on RNAs from these cell lines to identify UPR target genes whose expression was dependent on ATF6α. Surprisingly, the induction of almost all UPR target genes by Tm treatment was largely unaffected by ATF6α depletion (Table 1). In Northern blot analysis, we confirmed that BiP, CHOP, Grp94, ERdj4, and p58IPK transcripts were not significantly decreased in iATF6α MEFs, a finding consistent with the gene array results (Fig. 4C and data not shown). These results imply either that ATF6α is minimally involved in the regulation of UPR genes or that there is a functional redundancy. If the latter explanation is correct, then one possibility is that either ATF6β or XBP-1 can compensate for its loss.
The recently described ATF6β, which heterodimerizes with ATF6α, is closely related structurally to ATF6α and is also a transmembrane ER protein (11, 66). Upon activation by stress it is processed to an active, soluble form that translocates to the nucleus and transactivates endogenous BiP expression and the UPRE reporter. We used the strategy above therefore to “knockdown” the expression of ATF6β in wt MEFs, as well as in ATF6α MEFs, to produce singly and doubly deficient cell lines. Northern (not shown) and Western (Fig. 4A, right panel) blot analysis revealed very reduced levels of ATF6β mRNA and protein in both cell lines. Surprisingly, the induction of UPR target genes BiP, CHOP, and Grp94, as assessed by Northern blot analysis, was normal not only in the singly ATF6β but also in the double iATF6α/β cell lines (Fig. 4C). Gene array analysis also revealed that UPR target gene expression in iATF6α/β cells largely resembled that in iATF6α cells (data not shown). This is consistent with the normal induction of the UPRE and ERSE reporters upon Tm treatment of iATF6β MEFs (Fig. 4B). We conclude that, in this system, neither ATF6α nor ATF6β is required for the induction of UPR target genes. The impaired activity of the UPRE and ERSE reporters in response to Tm in iATF6α and double iATF6α/β MEFs (Fig. 4B), however, suggests that there exist additional UPR target genes, regulated by ATF6α, that we have not yet identified. Further, we have noted that XBP-1 and ATF6α synergistically transactivate the UPRE reporter (not shown).
UPR target gene expression in MEFs lacking both XBP-1 and ATF6α largely resembles that in single XBP-1-deficient MEFs.
We sought to assess whether there was functional redundancy between XBP-1 and ATF6α. We therefore transduced XBP-1−/− MEFs with the ATF6α RNA polymerase III-driven siRNA expression plasmid to generate MEFs that were doubly deficient in both XBP-1 and ATF6α, as shown by Western blot analysis (Fig. 5A). Gene array (not shown) and Northern blot analysis on RNA harvested from this MEF cell line in the presence or absence of Tm revealed that induction of the majority of UPR target genes that did not require XBP-1 was still only minimally decreased in the absence of both XBP-1 and ATF6α (Fig. 5B). BiP induction was found to be slightly more impaired in XBP-1/ATF6α doubly deficient cells than in either singly deficient cell line. However, the induction of Armet and Grp94 by Tm, only modestly diminished in the XBP-1 or ATF6α singly deficient MEFs, was significantly suppressed in the doubly deficient MEFs (Fig. 5B). These experiments make two important points. First, neither XBP-1 nor ATF6α can account for the induction of several of the prototypical ER stress response genes such as BiP. Second, Armet and Grp94 are examples of chaperone genes whose expression requires either, but not both, ATF6α or XBP-1. These doubly deficient cell lines should prove to be valuable reagents in searching for additional novel factors that can control target gene induction during the UPR.
FIG. 5.
UPR target gene expression in cells that lack both XBP-1 and ATF6α. (A) An XBP-1−/−/ATF6α doubly deficient MEF cell line was generated by transferring siRNA for ATF6α into XBP-1−/− MEF cells. ATF6α protein was absent in the doubly deficient cells as confirmed by Western blot analysis with anti-ATF6α antibody. (B) Total RNA was isolated from the indicated cell lines that were untreated or treated with Tm for 6 h and subjected to Northern blot analysis. The same blot was hybridized sequentially with BiP, CHOP, Armet, ERdj4, p58IPK, and Grp94 probes. Ethidium bromide staining of the gel before blotting is shown at the bottom as a loading control.
Dominant-negative XBP-1 suppresses UPR gene induction.
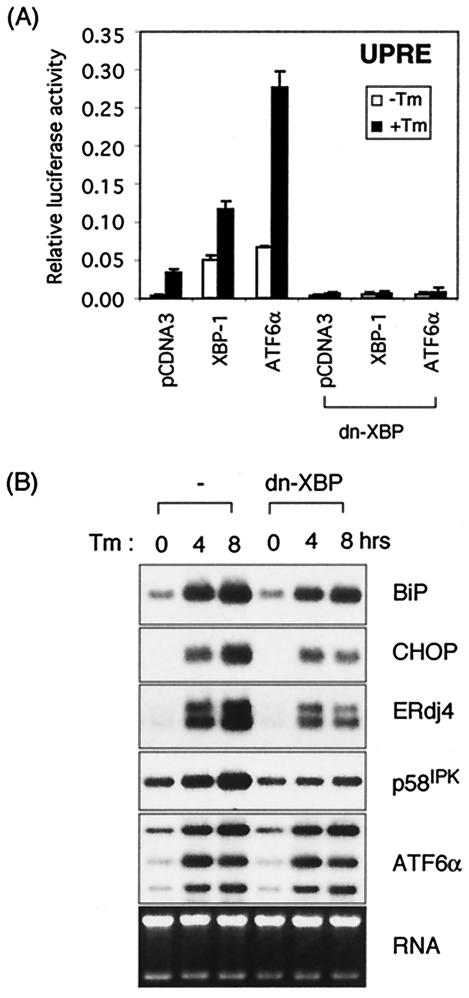
Since the N-terminal half (amino acids 1 to 188) of the XBP-1s protein lacks a transactivation domain but retains the leucine zipper motif essential for DNA binding and dimerization, it might function as a dominant negative that would physically interact with not only XBP-1 and ATF6α (unpublished observations) but also with other putative factors that associated with it. We tested the function of this mutant in reporter assays and found that dn-XBP completely abolished transactivation of the UPRE reporter by XBP-1s (Fig. 6A). Similarly, dn-XBP inhibited the transactivation of the UPRE reporter by ATF6α, demonstrating that dn-XBP dimerizes with both XBP-1 and with ATF6α. We generated a stable cell line overexpressing dn-XBP to inhibit the function of both XBP-1 and ATF6α and examined its effect on endogenous UPR target gene expression. dn-XBP significantly suppressed the induction of XBP-1 target genes, ERdj4 and p58IPK (Fig. 6B), although it did not completely inhibit XBP-1 and ATF6α activity, as evidenced by the residual ERdj4 and p58IPK expression in dn-XBP as opposed to XBP-1−/− MEFs. Interestingly, it also suppressed the induction of CHOP (Fig. 6B). Considering that CHOP induction was not significantly influenced by either XBP-1 or ATF6α or ATF6β loss singly or doubly, we conclude that CHOP induction requires another leucine zipper transcription factor that associates with dn-XBP. Thus, the results obtained with dn-XBP provide additional information not apparent in the analysis of the deficient MEFs above.
FIG. 6.
Dominant negative XBP-1 suppresses both XBP-1 and ATF6α activity. (A) The 5xATF6GL3 reporter plasmid was cotransfected with either pCGNATF6α or XBP-u/s plasmids into MEF cells with or without the dominant-negative XBP-1 expression plasmid. A total of 100 ng of DNA was used for each transfection except for pCDNA3.1, which was added to give 1 μg of DNA in total. Luciferase assays were performed as described in the legend to Fig. 4. (B) MEF and MEF-dn-XBP cells that stably express dominant-negative XBP-1 protein were treated with 10 μg of Tm/ml for the indicated time periods. Total RNAs were isolated and subjected to Northern blot analysis. The same blot was hybridized sequentially with BiP, CHOP, ERdj4, p58IPK, and ATF6α probes. Ethidium bromide staining of the gel before blotting is shown at the bottom as a loading control.
DISCUSSION
The UPR ensures the efficient translocation of newly synthesized peptides across the ER membrane and their subsequent folding, maturation, and transport by activating the expression of chaperone genes (16, 17). Two of the signaling systems that control the UPR are the IRE1/XBP-1 and ATF6 pathways. The relationship between XBP-1 and ATF6, two members of the basic region/leucine zipper class of transcription factors, has been unclear. Here we used DNA microarray analysis to search for genes regulated by XBP-1 and by ATF6α/β. Gene expression in MEFs derived from XBP-1-deficient embryos was compared to that in wt MEFs in the presence or absence of Tm, an agent that evokes the UPR. Similar analyses were carried out in ATF6α- and ATF6β-deficient MEFs generated by RNAi and in MEFs deficient for both XBP-1 and ATF6α. Two major conclusions emerged from this analysis. First, the expression of only a subset of UPR target genes depends on XBP-1. Second, we found that, in contrast to previously published work, neither ATF6α nor β were essential for the expression of a majority of UPR target genes, including BiP.
In the present study, we have identified a series of XBP-1-dependent UPR target genes: ERdj4, p58IPK, EDEM, RAMP-4, PDI-P5, and HEDJ, all of which appear to act in the ER. ERdj4 (46), p58IPK (29), and HEDJ (67) are localized to the ER and display Hsp40-like ATPase augmenting activity for the Hsp70 family chaperone proteins. EDEM was shown to be critically involved in the ERAD pathway by facilitating the degradation of ERAD substrates (13, 30, 31, 63). RAMP4 is a recently identified protein implicated in glycosylation and stabilization of membrane proteins in response to stress (42, 56, 59). PDI-P5 has homology to protein disulfide isomerase, which is thought to be involved in disulfide bond formation (18). Collectively, these results suggest that the IRE1/XBP-1 pathway is required for efficient protein folding, maturation, and degradation in the ER. Thus, it is not necessarily surprising that XBP-1 controls only a subset of UPR target genes.
Analysis of XBP-1−/−/iATF6α MEF cells revealed uncompromised induction of some UPR target genes upon ER stress. The PERK/ATF4 pathway is clearly responsible for the activation of some of these genes (10). This third UPR signaling pathway is activated by the PERK protein kinase. PERK phosphorylates eIF2α, which induces a transient suppression of protein translation, accompanied by induction of transcription factor(s) such as ATF4 (8). eIF2α is also phosphorylated under various cellular stress conditions by specific kinases, double-strand RNA activated protein kinase PKR, the amino acid control kinase GCN2, and the heme-regulated inhibitor HRI (17, 39). Since genes that are induced by the PERK pathway are also induced by other stress signals, such as amino acid deprivation, it is likely that PERK-dependent UPR target genes carry out common cellular defense mechanisms, such as cellular homeostasis, apoptosis, and cell cycle (10). Collectively, we propose that ER stress activates IRE/XBP-1 and PERK/eIF2α pathways to ensure proper maturation and degradation of secretory proteins and to effect common cellular defense mechanisms, respectively.
The reliance of p58IPK gene expression on XBP-1 is exciting since it connects two of the UPR signaling pathways, IRE1/XBP-1 and PERK. P58IPK was originally identified as a 58-kDa inhibitor of PKR in influenza virus-infected kidney cells (24) and described to downregulate the activity of PKR by binding to its kinase domain (15). It also has a J domain in the C terminus that has been shown to participate in interactions with Hsp70 family proteins (29). Recently, Katze and coworkers demonstrated that p58IPK (i) interacts with PERK, which is structurally similar to PKR, (ii) inhibits its eIF2α kinase activity, and (iii) is induced during the UPR by virtue of an ER stress response element in its promoter region (60). Our data suggest that XBP-1 is the transcription factor that controls p58IPK expression during the UPR. This has functional consequences since upregulation of p58IPK upon ER stress may relieve eIF2α phosphorylation and the subsequent protein translation induced by PERK in a negative-feedback manner.
Ohtsuka and Hata reported 23 mouse and human Hsp40/DnaJ homologs which were known genes or novel EST clones found in the DDBJ/GenBank/EMBL DNA database (32). Potential subcellular localization sites of these Hsp40 members were predicted based on their amino acid sequences and indicated that p58IPK, ERdj4, and Mtj1 likely have transmembrane domains and may be localized preferentially in the ER. Indeed, these proteins were confirmed to be localized to the ER, where they facilitate the ATPase activity of the Hsp70 chaperones (21, 46, 60). Recently, two additional ER Hsp40 members, hSec63 (49) and HEDJ (6), have been identified. XBP-1-dependent induction of p58IPK, ERdj4, and HEDJ suggests that an important role of XBP-1 in the UPR is to control the expression of some cochaperones that activate ER resident Hsp70 proteins. Mice that lack XBP-1 die in utero from liver hypoplasia (34), while mice lacking XBP-1 in the lymphoid system fail to generate plasma cells and hence antibodies (35, 36). The absolute dependence of these genes, i.e., ERdj4 and p58IPK, EDEM, Ramp4, PDI-P5, HEDJ, and others still unknown, on XBP-1 for expression suggests that they have an important function in the UPR in plasma cells.
XBP-1 and ATF6α have both been implicated in the function of the UPR. It has been shown by others that ATF6 is involved in the induction of a subset of UPR target genes (61, 64, 65, 68) and that ATF6α(1-373) was sufficient for the induction of several UPR target genes, including BiP and CHOP (33). Thus, our failure to uncover UPR target genes regulated by ATF6 was unexpected, especially since we found that the activity of the UPRE and ERSE reporters was completely absent or significantly diminished in ATF6α knockdown cells. It was puzzling that the induction of BiP mRNA was only marginally reduced in the absence of either ATF6α, ATF6β, or both, given that BiP induction was completely abolished in S2P-deficient CHO cells that failed to process ATF6α and presumably ATF6β as well (23, 61). A trivial explanation is the presence of residual ATF6α in the iATF6α MEFs. However, our failure to detect any processed ATF6α protein, coupled with the inhibition of ATF6α-dependent UPRE and ERSE activity, in this cell line makes this somewhat unlikely. This discrepancy may be due to differences in the cell type studied. One explanation is that XBP-1 may compensate for ATF6α, especially since they share similar DNA-binding specificities (unpublished observations). Alternatively, an intriguing possibility is that there is an additional UPR transcription factor that is activated through proteolysis by S2P similar to ATF6α and ATF6β. It is also certainly possible that there are ATF6α-specific UPR target genes that were not identified in our analysis, a scenario that is suggested by our observation that XBP-1 and ATF6α can synergistically activate the UPRE.
It has been proposed that XBP-1 is situated downstream of ATF6α as an explanation for the observation that ATF6α transactivates but in vitro-translated ATF6α fails to bind the UPRE reporter (64, 65). Although it has been reported that ATF6α transactivates the XBP-1 promoter, we found no evidence for the regulation of endogenous XBP-1 by ATF6α, since XBP-1 mRNA was normally induced in iATF6α cells. On the contrary, our data suggested that ATF6α was situated downstream of XBP-1 since the induction of mouse ATF6α mRNA upon ER stress was partially compromised in the absence of XBP-1. However, given that the induction of ATF6α by XBP-1 is modest and that ATF6α is primarily regulated by posttranslational mechanisms, we suggest that these two factors are situated largely in parallel pathways.
ATF6β is structurally related to ATF6α, with highest similarity in the b-zip domain (11, 66). Both ATF6α and ATF6β are proteolytically processed upon ER stress to release the N-terminal transactivator fragment. Interestingly, however, we found no evidence for a requirement of ATF6β for UPRE or ERSE reporter activity or for the induction of UPR target genes. It remains to be determined whether ATF6β is completely redundant with XBP-1 and ATF6α or has a specific, yet-to-be-determined role in UPR target gene expression.
The results obtained with reporter assays versus endogenous gene expression of certain UPR target genes such as BiP deserve comment. ERSE and UPRE motifs are extensively characterized DNA sequences that are responsive to ER stress (37, 57). UPRE was first identified as an artificial consensus DNA sequence that bound recombinant ATF6α protein (57) and independently was found to be strikingly similar to the optimal XBP-1 binding sequences (4). In contrast to the UPRE, whose motif has yet to be identified in the authentic promoter of any endogenous genes, ERSE CCAAT(N)9CCACG sequences (with N being a GC or GA rich region of 9 bp) is frequently found in the promoter region of well-known UPR target genes such as BiP, grp94, and CHOP (37, 54). It has been shown that the CCAAT motif is occupied by the constitutive transcription factor NF-Y and that the CCACG region is responsible for the inducible expression observed upon ER stress (37, 54). Both XBP-1 and ATF6 bind to the CCACG motif only in the presence of NF-Y (64).
We have shown that both XBP-1 and ATF6α regulate ERSE and UPRE reporters and that both BiP and CHOP promoters also failed to be induced in XBP-1/ATF6α double deficient cells by Tm (unpublished observations). However, neither XBP-1 nor ATF6 is a significant regulator of endogenous BiP expression. We conclude that there is an additional cis-acting element in the BiP and CHOP promoters that is responsible for their ER stress-induced expression. In contrast, the reporter gene assays, taken together with endogenous gene expression of certain UPR genes such as Armet and Grp94, suggested that XBP-1 and ATF6 might compensate for each other. The promoter region of the mouse Grp94 and Armet genes has three well-conserved ERSE motifs and one ERSE-II motif, respectively (unpublished observations). ERSE-II is similar to the ERSE motif, which consists of CCAAT and CCACG motifs separated by a single nucleotide in opposite directions (19). Thus, the ERSE and the ERSE-II motifs may be the critical control elements for Grp94 and Armet but not for BiP and CHOP. Rather, PERK-dependent transcription factors (i.e., ATF4) may control the induction of CHOP and a series of UPR target genes implicated in amino acid metabolism (8, 10). It was proposed that PERK activates unidentified transcription factor(s) in addition to ATF4, since only a subset of PERK-dependent UPR target genes was affected in ATF4-deficient cells (10). Deletion of XBP-1, ATF6, and PERK in various combinations will be necessary to establish the roles of these UPR signaling pathways.
Acknowledgments
This work was supported by NIH grant AI32412 (L.H.G.), an award from the Multiple Myeloma Research Foundation (L.H.G.), an award from the Ellison Medical Foundation, and an Irvington Institute postdoctoral fellowship award (N.N.I.).
We thank K. Mori for ATF6α and ATF6β antibodies; R. Prywes for the pCGNATF6 and pCGNATF6(1-373) constructs; D. Ron for the IRE1α−/− MEFs; R. J. Kaufman for the Grp94 and BiP probes; L. Hendershot for the HEDJ probe; K. Mowen, M. Oukka, and M. Grusby for thoughtful comments on the manuscript; and C. McCall for help with manuscript preparation.
REFERENCES
- 1.Bukau, B., and A. L. Horwich. 1998. The Hsp70 and Hsp60 chaperone machines. Cell 92:351-366. [DOI] [PubMed] [Google Scholar]
- 2.Calfon, M., H. Zeng, F. Urano, J. H. Till, S. R. Hubbard, H. P. Harding, S. G. Clark, and D. Ron. 2002. IRE1 couples endoplasmic reticulum load to secretory capacity by processing the XBP-1 mRNA. Nature 415:92-96. [DOI] [PubMed] [Google Scholar]
- 3.Chevalier, M., H. Rhee, E. C. Elguindi, and S. Y. Blond. 2000. Interaction of murine BiP/GRP78 with the DnaJ homologue MTJ1. J. Biol. Chem. 275:19620-19627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Clauss, I. M., M. Chu, J.-L. Zhao, and L. H. Glimcher. 1996. The basic domain/leucine zipper protein hXBP-1 preferentially binds to and transactivates CRE-like sequences containing an ACGT core. Nucleic Acids Res. 24:1855-1864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cox, J. S., and P. Walter. 1996. A novel mechanism for regulating activity of a transcription factor that controls the unfolded protein response. Cell 87:391-404. [DOI] [PubMed] [Google Scholar]
- 6.Dudek, J., J. Volkmer, C. Bies, S. Guth, A. Muller, M. Lerner, P. Feick, K. H. Schafer, E. Morgenstern, F. Hennessy, G. L. Blatch, K. Janoscheck, N. Heim, P. Scholtes, M. Frien, W. Nastainczyk, and R. Zimmermann. 2002. A novel type of co-chaperone mediates transmembrane recruitment of DnaK-like chaperones to ribosomes. EMBO J. 21:2958-2967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Foti, D. M., A. Welihinda, R. J. Kaufman, and A. S. Lee. 1999. Conservation and divergence of the yeast and mammalian unfolded protein response. Activation of specific mammalian endoplasmic reticulum stress element of the grp78/BiP promoter by yeast Hac1. J. Biol. Chem. 274:30402-30409. [DOI] [PubMed] [Google Scholar]
- 8.Harding, H. P., I. Novoa, Y. Zhang, H. Zeng, R. Wek, M. Schapira, and D. Ron. 2000. Regulated translation initiation controls stress-induced gene expression in mammalian cells. Mol. Cell 6:1099-1108. [DOI] [PubMed] [Google Scholar]
- 9.Harding, H. P., Y. Zhang, and D. Ron. 1999. Protein translation and folding are coupled by an endoplasmic-reticulum-resident kinase. Nature 397:271-274. [DOI] [PubMed] [Google Scholar]
- 10.Harding, H. P., Y. Zhang, H. Zeng, I. Novoa, P. D. Lu, M. Calfon, N. Sadri, C. Yun, B. Popko, R. Paules, D. F. Stojdl, J. C. Bell, T. Hettmann, J. M. Leiden, and D. Ron. 2003. An integrated stress response regulates amino Acid metabolism and resistance to oxidative stress. Mol. Cell 11:619-633. [DOI] [PubMed] [Google Scholar]
- 11.Haze, K., T. Okada, H. Yoshida, H. Yanagi, T. Yura, M. Negishi, and K. Mori. 2001. Identification of the G13 (cAMP-response-element-binding protein-related protein) gene product related to activating transcription factor 6 as a transcriptional activator of the mammalian unfolded protein response. Biochem. J. 355:19-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Haze, K., H. Yoshida, H. Yanagi, T. Yura, and K. Mori. 1999. Mammalian transcription factor ATF6 is synthesized as a transmembrane protein and activated by proteolysis in response to endoplasmic reticulum stress. Mol. Biol. Cell 10:3787-3799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hosokawa, N., I. Wada, K. Hasegawa, T. Yorihuzi, L. O. Tremblay, A. Herscovics, and K. Nagata. 2001. A novel ER alpha-mannosidase-like protein accelerates ER-associated degradation. EMBO Rep. 2:415-422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Iwakoshi, N. N., A. H. Lee, P. Vallabhajosyula, K. L. Otipoby, K. Rajewsky, and L. H. Glimcher. 2003. Plasma cell differentiation and the unfolded protein response intersect the transcription factor XBP-1. Nat. Immunol. 4:321-329. [DOI] [PubMed] [Google Scholar]
- 15.Katze, M. G. 1995. Regulation of the interferon-induced PKR: can viruses cope? Trends Microbiol. 3:75-78. [DOI] [PubMed] [Google Scholar]
- 16.Kaufman, R. J. 2002. Orchestrating the unfolded protein response in health and disease. J. Clin. Investig. 110:1389-1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kaufman, R. J. 1999. Stress signaling from the lumen of the endoplasmic reticulum: coordination of gene transcriptional and translational controls. Genes Dev. 13:1211-1233. [DOI] [PubMed] [Google Scholar]
- 18.Kikuchi, M., E. Doi, I. Tsujimoto, T. Horibe, and Y. Tsujimoto. 2002. Functional analysis of human P5, a protein disulfide isomerase homologue. J. Biochem. 132:451-455. [DOI] [PubMed] [Google Scholar]
- 19.Kokame, K., H. Kato, and T. Miyata. 2001. Identification of ERSE-II, a new cis-acting element responsible for the ATF6-dependent mammalian unfolded protein response. J. Biol. Chem. 276:9199-9205. [DOI] [PubMed] [Google Scholar]
- 20.Korth, M. J., C. N. Lyons, M. Wambach, and M. G. Katze. 1996. Cloning, expression and cellular localization of the oncogenic 58-kDa inhibitor of the RNA-activated human and mouse protein kinase. Gene 170:181-188. [DOI] [PubMed] [Google Scholar]
- 21.Kurisu, J., A. Honma, H. Miyajima, S. Kondo, M. Okumura, and K. Imaizumi. 2003. MDG1/ERdj4, an ER-resident DnaJ family member, suppresses cell death induced by ER stress. Genes Cells 8:189-202. [DOI] [PubMed] [Google Scholar]
- 22.Lee, A. H., J. H. Hong, and Y. S. Seo. 2000. Tumor necrosis factor-alpha and interferon-gamma synergistically activate the RANTES promoter through nuclear factor κB and interferon regulatory factor 1 (IRF-1) transcription factors. Biochem. J. 350(Pt. 1):131-138. [PMC free article] [PubMed] [Google Scholar]
- 23.Lee, K., W. Tirasophon, X. Shen, M. Michalak, R. Prywes, T. Okada, H. Yoshida, K. Mori, and R. J. Kaufman. 2002. IRE1-mediated unconventional mRNA splicing and S2P-mediated ATF6 cleavage merge to regulate XBP1 in signaling the unfolded protein response. Genes Dev. 16:452-466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee, T. G., J. Tomita, A. G. Hovanessian, and M. G. Katze. 1990. Purification and partial characterization of a cellular inhibitor of the interferon-induced protein kinase of Mr 68,000 from influenza virus-infected cells. Proc. Natl. Acad. Sci. USA 87:6208-6212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li, M., P. Baumeister, B. Roy, T. Phan, D. Foti, S. Luo, and A. S. Lee. 2000. ATF6 as a transcription activator of the endoplasmic reticulum stress element: thapsigargin stress-induced changes and synergistic interactions with NF-Y and YY1. Mol. Cell. Biol. 20:5096-5106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lindemann, D., E. Patriquin, S. Feng, and R. C. Mulligan. 1997. Versatile retrovirus vector systems for regulated gene expression in vitro and in vivo. Mol. Med. 3:466-476. [PMC free article] [PubMed] [Google Scholar]
- 27.Luo, G., P. Ducy, M. D. McKee, G. J. Pinero, E. Loyer, R. R. Behringer, and G. Karsenty. 1997. Spontaneous calcification of arteries and cartilage in mice lacking matrix GLA protein. Nature 386:78-81. [DOI] [PubMed] [Google Scholar]
- 28.Ma, Y., and L. M. Hendershot. 2001. The unfolding tale of the unfolded protein response. Cell 107:827-830. [DOI] [PubMed] [Google Scholar]
- 29.Melville, M. W., S. L. Tan, M. Wambach, J. Song, R. I. Morimoto, and M. G. Katze. 1999. The cellular inhibitor of the PKR protein kinase, P58(IPK), is an influenza virus-activated co-chaperone that modulates heat shock protein 70 activity. J. Biol. Chem. 274:3797-3803. [DOI] [PubMed] [Google Scholar]
- 30.Molinari, M., V. Calanca, C. Galli, P. Lucca, and P. Paganetti. 2003. Role of EDEM in the release of misfolded glycoproteins from the calnexin cycle. Science 299:1397-1400. [DOI] [PubMed] [Google Scholar]
- 31.Oda, Y., N. Hosokawa, I. Wada, and K. Nagata. 2003. EDEM as an acceptor of terminally misfolded glycoproteins released from calnexin. Science 299:1394-1397. [DOI] [PubMed] [Google Scholar]
- 32.Ohtsuka, K., and M. Hata. 2000. Mammalian HSP40/DNAJ homologs: cloning of novel cDNAs and a proposal for their classification and nomenclature. Cell Stress Chaperones 5:98-112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Okada, T., H. Yoshida, R. Akazawa, M. Negishi, and K. Mori. 2002. Distinct roles of activating transcription factor 6 (ATF6) and double-stranded RNA-activated protein kinase-like endoplasmic reticulum kinase (PERK) in transcription during the mammalian unfolded protein response. Biochem. J. 366:585-594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Reimold, A. M., A. Etkin, I. Clauss, A. Perkins, D. S. Friend, J. Zhang, H. F. Horton, A. Scott, S. H. Orkin, M. C. Byrne, M. J. Grusby, and L. H. Glimcher. 2000. An essential role in liver development for transcription factor XBP-1. Genes Dev. 14:152-157. [PMC free article] [PubMed] [Google Scholar]
- 35.Reimold, A. M., N. N. Iwakoshi, J. Manis, P. Vallabhajosyula, E. Szomolanyi-Tsuda, E. M. Gravallese, D. Friend, M. J. Grusby, F. Alt, and L. H. Glimcher. 2001. Plasma cell differentiation requires transcription factor XBP-1. Nature 412:300-307. [DOI] [PubMed] [Google Scholar]
- 36.Reimold, A. M., P. D. Ponath, Y. S. Li, R. R. Hardy, C. S. David, J. L. Strominger, and L. H. Glimcher. 1996. Transcription factor B-cell lineage-specific activator protein regulates the gene for human X-box binding protein 1. J. Exp. Med. 183:393-401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Roy, B., and A. S. Lee. 1999. The mammalian endoplasmic reticulum stress response element consists of an evolutionarily conserved tripartite structure and interacts with a novel stress-inducible complex. Nucleic Acids Res. 27:1437-1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ruegsegger, U., J. H. Leber, and P. Walter. 2001. Block of HAC1 mRNA translation by long-range base pairing is released by cytoplasmic splicing upon induction of the unfolded protein response. Cell 107:103-114. [DOI] [PubMed] [Google Scholar]
- 39.Samuel, C. E. 1993. The eIF-2 alpha protein kinases, regulators of translation in eukaryotes from yeasts to humans. J. Biol. Chem. 268:7603-7606. [PubMed] [Google Scholar]
- 40.Scheuner, D., B. Song, E. McEwen, C. Liu, R. Laybutt, P. Gillespie, T. Saunders, S. Bonner-Weir, and R. J. Kaufman. 2001. Translational control is required for the unfolded protein response and in vivo glucose homeostasis. Mol. Cell 7:1165-1176. [DOI] [PubMed] [Google Scholar]
- 41.Schmid, D., A. Baici, H. Gehring, and P. Christen. 1994. Kinetics of molecular chaperone action. Science 263:971-973. [DOI] [PubMed] [Google Scholar]
- 42.Schroder, K., B. Martoglio, M. Hofmann, C. Holscher, E. Hartmann, S. Prehn, T. A. Rapoport, and B. Dobberstein. 1999. Control of glycosylation of MHC class II-associated invariant chain by translocon-associated RAMP4. EMBO J. 18:4804-4815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shamu, C. E., and P. Walter. 1996. Oligomerization and phosphorylation of the Ire1p kinase during intracellular signaling from the endoplasmic reticulum to the nucleus. EMBO J. 15:3028-3039. [PMC free article] [PubMed] [Google Scholar]
- 44.Shen, J., X. Chen, L. Hendershot, and R. Prywes. 2002. ER stress regulation of ATF6 localization by dissociation of BiP/GRP78 binding and unmasking of Golgi localization signals. Dev. Cell 3:99-111. [DOI] [PubMed] [Google Scholar]
- 45.Shen, X., R. E. Ellis, K. Lee, C. Y. Liu, K. Yang, A. Solomon, H. Yoshida, R. Morimoto, D. M. Kurnit, K. Mori, and R. J. Kaufman. 2001. Complementary signaling pathways regulate the unfolded protein response and are required for Caenorhabditis elegans development. Cell 107:893-903. [DOI] [PubMed] [Google Scholar]
- 46.Shen, Y., L. Meunier, and L. M. Hendershot. 2003. Identification and characterization of a novel endoplasmic reticulum (ER) DnaJ homologue, which stimulates ATPase activity of BiP in vitro and is induced by ER stress. J. Biol. Chem. 277:15947-15956. [DOI] [PubMed] [Google Scholar]
- 47.Shi, Y., K. M. Vattem, R. Sood, J. An, J. Liang, L. Stramm, and R. C. Wek. 1998. Identification and characterization of pancreatic eukaryotic initiation factor 2 alpha-subunit kinase, PEK, involved in translational control. Mol. Cell. Biol. 18:7499-7509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sidrauski, C., J. S. Cox, and P. Walter. 1996. tRNA ligase is required for regulated mRNA splicing in the unfolded protein response. Cell 87:405-413. [DOI] [PubMed] [Google Scholar]
- 49.Skowronski, J., C. Jolicoeur, S. Alpert, and D. Hanahan. 1990. Determinants of the B-cell response against a transgenic autoantigen. Proc. Natl. Acad. Sci. USA 87:7487-7491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sui, G., C. Soohoo, B. Affar el, F. Gay, Y. Shi, and W. C. Forrester. 2002. A DNA vector-based RNAi technology to suppress gene expression in mammalian cells. Proc. Natl. Acad. Sci. USA 99:5515-5520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tirasophon, W., K. Lee, B. Callaghan, A. Welihinda, and R. J. Kaufman. 2000. The endoribonuclease activity of mammalian IRE1 autoregulates its mRNA and is required for the unfolded protein response. Genes Dev. 14:2725-2736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tirasophon, W., A. A. Welihinda, and R. J. Kaufman. 1998. A stress response pathway from the endoplasmic reticulum to the nucleus requires a novel bifunctional protein kinase/endoribonuclease (Ire1p) in mammalian cells. Genes Dev. 12:1812-1824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Travers, K. J., C. K. Patil, L. Wodicka, D. J. Lockhart, J. S. Weissman, and P. Walter. 2000. Functional and genomic analyses reveal an essential coordination between the unfolded protein response and ER-associated degradation. Cell 101:249-258. [DOI] [PubMed] [Google Scholar]
- 54.Ubeda, M., and J. F. Habener. 2000. CHOP gene expression in response to endoplasmic-reticular stress requires NFY interaction with different domains of a conserved DNA-binding element. Nucleic Acids Res. 28:4987-4997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Urano, F., A. Bertolotti, and D. Ron. 2000. IRE1 and efferent signaling from the endoplasmic reticulum. J. Cell Sci. 113:3697-3702. [DOI] [PubMed] [Google Scholar]
- 56.Wang, L., and B. Dobberstein. 1999. Oligomeric complexes involved in translocation of proteins across the membrane of the endoplasmic reticulum. FEBS Lett. 457:316-322. [DOI] [PubMed] [Google Scholar]
- 57.Wang, Y., J. Shen, N. Arenzana, W. Tirasophon, R. J. Kaufman, and R. Prywes. 2000. Activation of ATF6 and an ATF6 DNA binding site by the endoplasmic reticulum stress response. J. Biol. Chem. 275:27013-27020. [DOI] [PubMed] [Google Scholar]
- 58.Welihinda, A. A., and R. J. Kaufman. 1996. The unfolded protein response pathway in Saccharomyces cerevisiae: oligomerization and trans-phosphorylation of Ire1p (Ern1p) are required for kinase activation. J. Biol. Chem. 271:18181-18187. [DOI] [PubMed] [Google Scholar]
- 59.Yamaguchi, A., O. Hori, D. M. Stern, E. Hartmann, S. Ogawa, and M. Tohyama. 1999. Stress-associated endoplasmic reticulum protein 1 (SERP1)/ribosome-associated membrane protein 4 (RAMP4) stabilizes membrane proteins during stress and facilitates subsequent glycosylation. J. Cell Biol. 147:1195-1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yan, W., C. L. Frank, M. J. Korth, B. L. Sopher, I. Novoa, D. Ron, and M. G. Katze. 2002. Control of PERK eIF2α kinase activity by the endoplasmic reticulum stress-induced molecular chaperone P58IPK. Proc. Natl. Acad. Sci. USA 99:15920-15925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ye, J., R. B. Rawson, R. Komuro, X. Chen, U. P. Dave, R. Prywes, M. S. Brown, and J. L. Goldstein. 2000. ER stress induces cleavage of membrane-bound ATF6 by the same proteases that process SREBPs. Mol. Cell 6:1355-1364. [DOI] [PubMed] [Google Scholar]
- 62.Yoshida, H., K. Haze, H. Yanagi, T. Yura, and K. Mori. 1998. Identification of the cis-acting endoplasmic reticulum stress response element responsible for transcriptional induction of mammalian glucose-regulated proteins. Involvement of basic leucine zipper transcription factors. J. Biol. Chem. 273:33741-33749. [DOI] [PubMed] [Google Scholar]
- 63.Yoshida, H., T. Matsui, N. Hosokawa, R. J. Kaufman, K. Nagata, and K. Mori. 2003. A time-dependent phase shift in the Mammalian unfolded protein response. Dev. Cell 4:265-271. [DOI] [PubMed] [Google Scholar]
- 64.Yoshida, H., T. Matsui, A. Yamamoto, T. Okada, and K. Mori. 2001. XBP1 mRNA is induced by ATF6 and spliced by IRE1 in response to ER stress to produce a highly active transcription factor. Cell 107:881-891. [DOI] [PubMed] [Google Scholar]
- 65.Yoshida, H., T. Okada, K. Haze, H. Yanagi, T. Yura, M. Negishi, and K. Mori. 2000. ATF6 activated by proteolysis binds in the presence of NF-Y (CBF) directly to the cis-acting element responsible for the mammalian unfolded protein response. Mol. Cell. Biol. 20:6755-6767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yoshida, H., T. Okada, K. Haze, H. Yanagi, T. Yura, M. Negishi, and K. Mori. 2001. Endoplasmic reticulum stress-induced formation of transcription factor complex ERSF including NF-Y (CBF) and activating transcription factors 6α and 6β that activates the mammalian unfolded protein response. Mol. Cell. Biol. 21:1239-1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yu, M., R. H. Haslam, and D. B. Haslam. 2000. HEDJ, an Hsp40 co-chaperone localized to the endoplasmic reticulum of human cells. J. Biol. Chem. 275:24984-24992. [DOI] [PubMed] [Google Scholar]
- 68.Zhu, C., F. E. Johansen, and R. Prywes. 1997. Interaction of ATF6 and serum response factor. Mol. Cell. Biol. 17:4957-4966. [DOI] [PMC free article] [PubMed] [Google Scholar]