Abstract
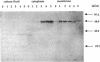
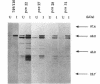
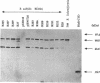
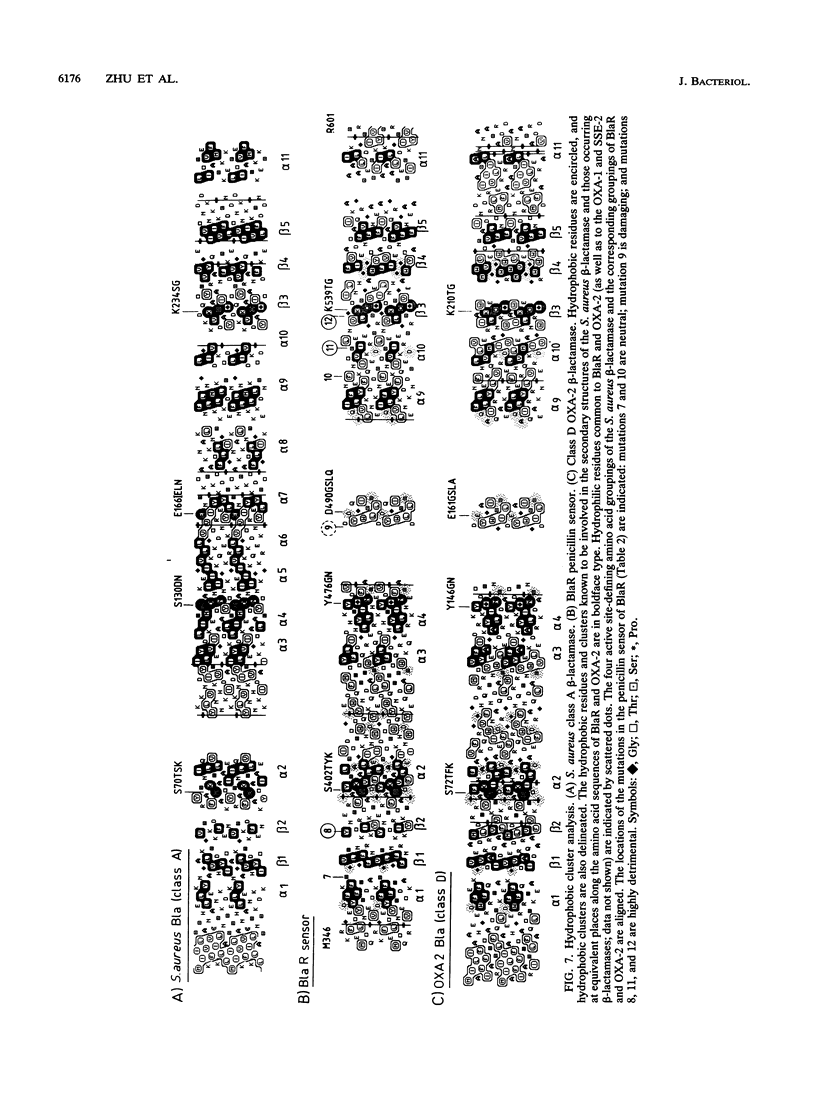
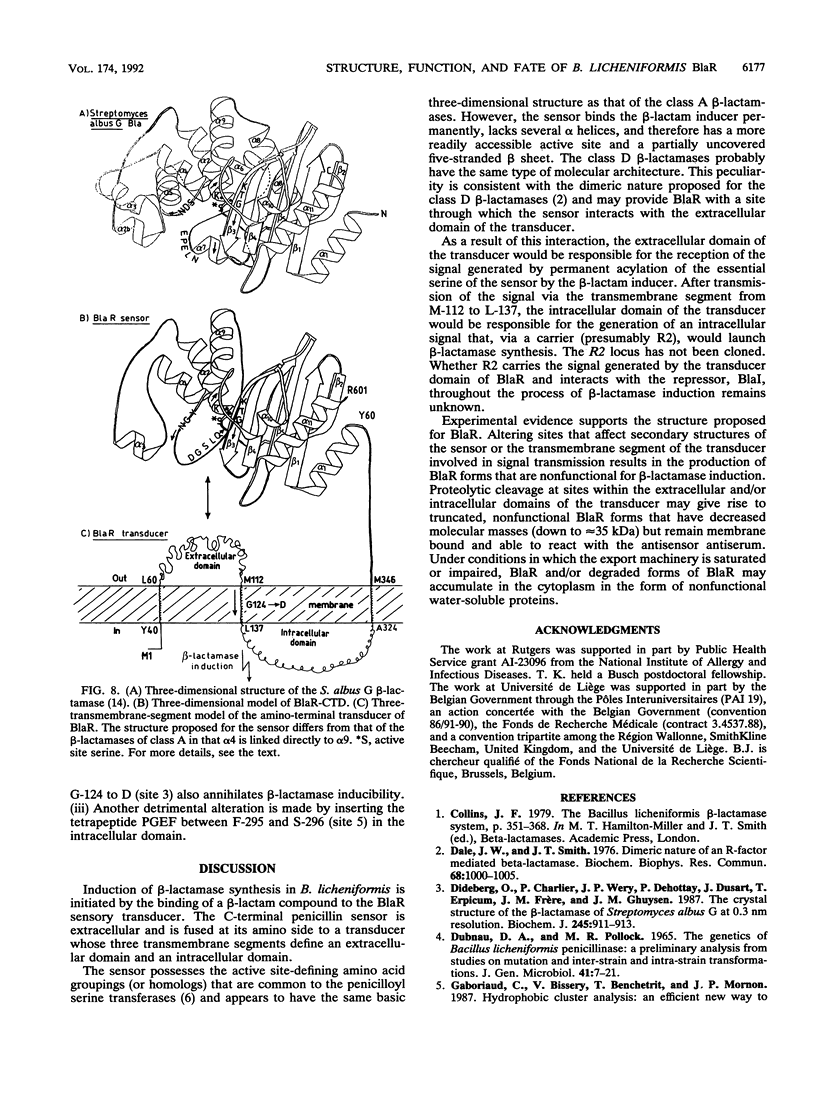
The membrane-spanning protein BlaR is essential for the induction of beta-lactamase in Bacillus licheniformis. Its nature and location were confirmed by the use of an antiserum specific for its carboxy-terminal penicillin sensor, its function was studied by genetic dissection, and the structure of the penicillin sensor was derived from hydrophobic cluster analysis of the amino acid sequence by using, as a reference, the class A beta-lactamases with known three-dimensional structures. During the first 2 h after the addition of the beta-lactam inducer, full-size BlaR, bound to the plasma membrane, is produced, and then beta-lactamase is produced. By 2 h after induction, BlaR is present in various (membrane-bound and cytosolic) forms, and there is a gradual decrease in beta-lactamase production. The penicillin sensors of BlaR and the class D beta-lactamases show strong similarities in primary structures. They appear to have the same basic spatial disposition of secondary structures as that of the class A beta-lactamases, except that they lack several alpha helices and, therefore, have a partially uncovered five-stranded beta sheet and a more readily accessible active site. Alterations of BlaR affecting conserved secondary structures of the penicillin sensor and specific sites of the transducer annihilate beta-lactamase inducibility.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Dale J. W., Smith J. T. The dimeric nature of an R-factor mediated beta-lactamase. Biochem Biophys Res Commun. 1976 Feb 9;68(3):1000–1005. doi: 10.1016/0006-291x(76)91245-6. [DOI] [PubMed] [Google Scholar]
- Dideberg O., Charlier P., Wéry J. P., Dehottay P., Dusart J., Erpicum T., Frère J. M., Ghuysen J. M. The crystal structure of the beta-lactamase of Streptomyces albus G at 0.3 nm resolution. Biochem J. 1987 Aug 1;245(3):911–913. doi: 10.1042/bj2450911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubnau D. A., Pollock M. R. The genetics of Bacillus licheniformis penicillinase: a preliminary analysis from studies on mutation and inter-strain and intra-strain transformations. J Gen Microbiol. 1965 Oct;41(1):7–21. doi: 10.1099/00221287-41-1-7. [DOI] [PubMed] [Google Scholar]
- Ghuysen J. M. Serine beta-lactamases and penicillin-binding proteins. Annu Rev Microbiol. 1991;45:37–67. doi: 10.1146/annurev.mi.45.100191.000345. [DOI] [PubMed] [Google Scholar]
- Henrissat B., Saloheimo M., Lavaitte S., Knowles J. K. Structural homology among the peroxidase enzyme family revealed by hydrophobic cluster analysis. Proteins. 1990;8(3):251–257. doi: 10.1002/prot.340080307. [DOI] [PubMed] [Google Scholar]
- Herzberg O. Refined crystal structure of beta-lactamase from Staphylococcus aureus PC1 at 2.0 A resolution. J Mol Biol. 1991 Feb 20;217(4):701–719. doi: 10.1016/0022-2836(91)90527-d. [DOI] [PubMed] [Google Scholar]
- Joris B., Ghuysen J. M., Dive G., Renard A., Dideberg O., Charlier P., Frère J. M., Kelly J. A., Boyington J. C., Moews P. C. The active-site-serine penicillin-recognizing enzymes as members of the Streptomyces R61 DD-peptidase family. Biochem J. 1988 Mar 1;250(2):313–324. doi: 10.1042/bj2500313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joris B., Ledent P., Dideberg O., Fonzé E., Lamotte-Brasseur J., Kelly J. A., Ghuysen J. M., Frère J. M. Comparison of the sequences of class A beta-lactamases and of the secondary structure elements of penicillin-recognizing proteins. Antimicrob Agents Chemother. 1991 Nov;35(11):2294–2301. doi: 10.1128/aac.35.11.2294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joris B., Ledent P., Kobayashi T., Lampen J. O., Ghuysen J. M. Expression in Escherichia coli of the carboxy terminal domain of the BLAR sensory-transducer protein of Bacillus licheniformis as a water-soluble Mr 26,000 penicillin-binding protein. FEMS Microbiol Lett. 1990 Jun 15;58(1):107–113. doi: 10.1016/0378-1097(90)90111-3. [DOI] [PubMed] [Google Scholar]
- Kobayashi T., Zhu Y. F., Nicholls N. J., Lampen J. O. A second regulatory gene, blaR1, encoding a potential penicillin-binding protein required for induction of beta-lactamase in Bacillus licheniformis. J Bacteriol. 1987 Sep;169(9):3873–3878. doi: 10.1128/jb.169.9.3873-3878.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamotte-Brasseur J., Dive G., Dideberg O., Charlier P., Frère J. M., Ghuysen J. M. Mechanism of acyl transfer by the class A serine beta-lactamase of Streptomyces albus G. Biochem J. 1991 Oct 1;279(Pt 1):213–221. doi: 10.1042/bj2790213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenzie T., Hoshino T., Tanaka T., Sueoka N. The nucleotide sequence of pUB110: some salient features in relation to replication and its regulation. Plasmid. 1986 Mar;15(2):93–103. doi: 10.1016/0147-619x(86)90046-6. [DOI] [PubMed] [Google Scholar]
- Oliver D. B., Beckwith J. Regulation of a membrane component required for protein secretion in Escherichia coli. Cell. 1982 Aug;30(1):311–319. doi: 10.1016/0092-8674(82)90037-x. [DOI] [PubMed] [Google Scholar]
- Salerno A. J., Lampen J. O. Differential transcription of the bla regulatory region during induction of beta-lactamase in Bacillus licheniformis. FEBS Lett. 1988 Jan 18;227(1):61–65. doi: 10.1016/0014-5793(88)81414-5. [DOI] [PubMed] [Google Scholar]
- Salerno A. J., Lampen J. O. Transcriptional analysis of beta-lactamase regulation in Bacillus licheniformis. J Bacteriol. 1986 Jun;166(3):769–778. doi: 10.1128/jb.166.3.769-778.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sargent M. G. Rapid fixed-time assay for penicillinase. J Bacteriol. 1968 Apr;95(4):1493–1494. doi: 10.1128/jb.95.4.1493-1494.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherratt D. J., Collins J. F. Analysis by transformation of the penicillinase system in Bacillus licheniformis. J Gen Microbiol. 1973 May;76(1):217–230. doi: 10.1099/00221287-76-1-217. [DOI] [PubMed] [Google Scholar]
- Smith P. K., Krohn R. I., Hermanson G. T., Mallia A. K., Gartner F. H., Provenzano M. D., Fujimoto E. K., Goeke N. M., Olson B. J., Klenk D. C. Measurement of protein using bicinchoninic acid. Anal Biochem. 1985 Oct;150(1):76–85. doi: 10.1016/0003-2697(85)90442-7. [DOI] [PubMed] [Google Scholar]
- Tabor S., Richardson C. C. A bacteriophage T7 RNA polymerase/promoter system for controlled exclusive expression of specific genes. Proc Natl Acad Sci U S A. 1985 Feb;82(4):1074–1078. doi: 10.1073/pnas.82.4.1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young R. A., Davis R. W. Efficient isolation of genes by using antibody probes. Proc Natl Acad Sci U S A. 1983 Mar;80(5):1194–1198. doi: 10.1073/pnas.80.5.1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y. F., Curran I. H., Joris B., Ghuysen J. M., Lampen J. O. Identification of BlaR, the signal transducer for beta-lactamase production in Bacillus licheniformis, as a penicillin-binding protein with strong homology to the OXA-2 beta-lactamase (class D) of Salmonella typhimurium. J Bacteriol. 1990 Feb;172(2):1137–1141. doi: 10.1128/jb.172.2.1137-1141.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]