Abstract
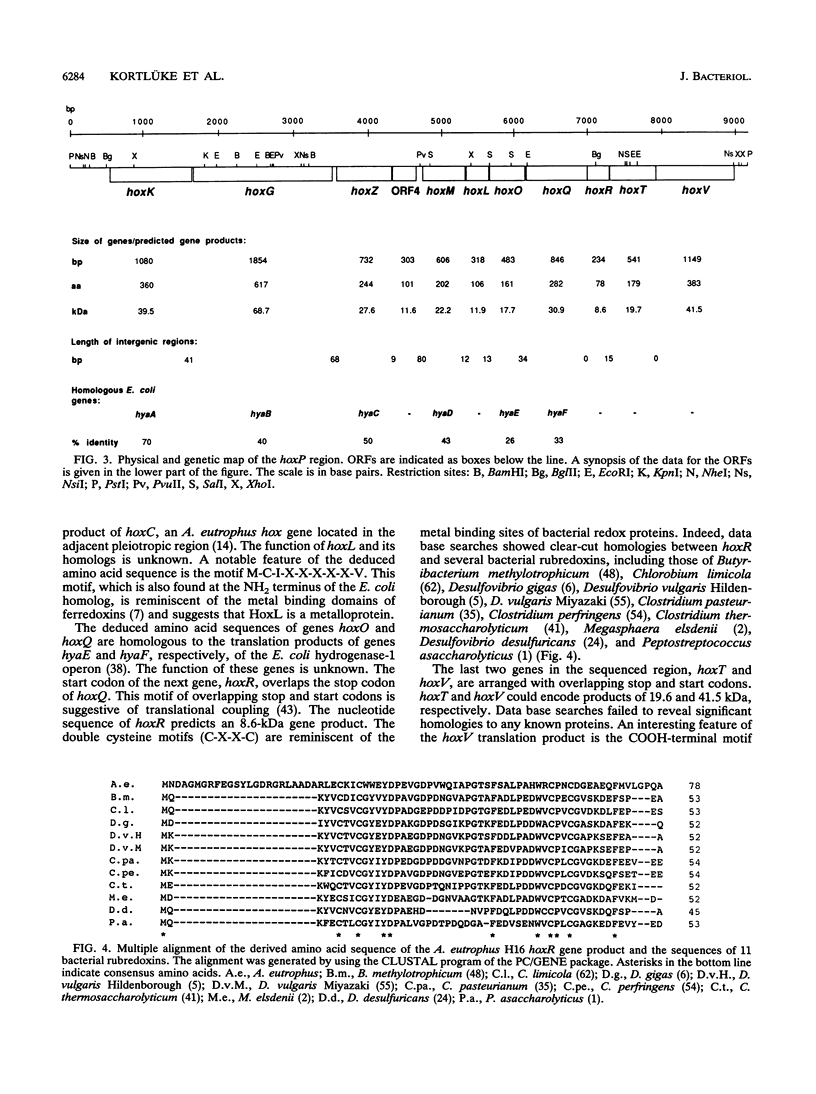
One of the key enzymes in the chemolithoautotrophic metabolism of Alcaligenes eutrophus H16 is a dimeric, membrane-associated hydrogenase. The genetic determinants of this enzyme are located on the endogenous megaplasmid pHG1 (G. Eberz, C. Hogrefe, C. Kortlüke, A. Kamienski, and B. Friedrich, J. Bacteriol. 168:636-641, 1986). Complementation studies showed that the information required for the formation of active membrane-bound hydrogenase occupies more than 7.5 kb of megaplasmid DNA. We cloned and sequenced this region and identified the genes encoding the two hydrogenase subunits (hoxK and hoxG). The nucleotide sequence contains nine additional closely spaced open reading frames. Immunoelectron microscopy showed that the gene product of one of these open reading frames (hoxM) is involved in the process leading to the attachment of hydrogenase to the membrane. Other open reading frames may encode additional processing functions and components of a hydrogenase-linked electron transport chain. Analysis of Tn5-B21-mediated transcriptional fusions provided evidence that the structural genes and accessory functions belong to at least three coordinately regulated transcriptional units.
Full text
PDF











Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bachmayer H., Benson A. M., Yasunobu K. T., Garrard W. T., Whiteley H. R. Nonheme iron proteins. IV. Structural studies of Micrococcus aerogenes rubredoxin. Biochemistry. 1968 Mar;7(3):986–996. doi: 10.1021/bi00843a016. [DOI] [PubMed] [Google Scholar]
- Bachmayer H., Yasunobu K. T., Peel J. L., Mayhew S. Non-heme iron proteins. V. The amino acid sequence of rubredoxin from Peptostreptococcus elsdenii. J Biol Chem. 1968 Mar 10;243(5):1022–1030. [PubMed] [Google Scholar]
- Bowien B., Schlegel H. G. Physiology and biochemistry of aerobic hydrogen-oxidizing bacteria. Annu Rev Microbiol. 1981;35:405–452. doi: 10.1146/annurev.mi.35.100181.002201. [DOI] [PubMed] [Google Scholar]
- Boyer H. W., Roulland-Dussoix D. A complementation analysis of the restriction and modification of DNA in Escherichia coli. J Mol Biol. 1969 May 14;41(3):459–472. doi: 10.1016/0022-2836(69)90288-5. [DOI] [PubMed] [Google Scholar]
- Brumlik M. J., Voordouw G. Analysis of the transcriptional unit encoding the genes for rubredoxin (rub) and a putative rubredoxin oxidoreductase (rbo) in Desulfovibrio vulgaris Hildenborough. J Bacteriol. 1989 Sep;171(9):4996–5004. doi: 10.1128/jb.171.9.4996-5004.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruschi M., Guerlesquin F. Structure, function and evolution of bacterial ferredoxins. FEMS Microbiol Rev. 1988 Apr-Jun;4(2):155–175. doi: 10.1111/j.1574-6968.1988.tb02741.x. [DOI] [PubMed] [Google Scholar]
- Bruschi M. The amino acid sequence of rubredoxin from the sulfate reducing bacterium, Desulfovibrio gigas. Biochem Biophys Res Commun. 1976 May 17;70(2):615–621. doi: 10.1016/0006-291x(76)91092-5. [DOI] [PubMed] [Google Scholar]
- Burke J. M., Breitenberger C., Heckman J. E., Dujon B., RajBhandary U. L. Cytochrome b gene of Neurospora crassa mitochondria. Partial sequence and location of introns at sites different from those in Saccharomyces cerevisiae and Aspergillus nidulans. J Biol Chem. 1984 Jan 10;259(1):504–511. [PubMed] [Google Scholar]
- Davison J., Heusterspreute M., Chevalier N., Ha-Thi V., Brunel F. Vectors with restriction site banks. V. pJRD215, a wide-host-range cosmid vector with multiple cloning sites. Gene. 1987;51(2-3):275–280. doi: 10.1016/0378-1119(87)90316-7. [DOI] [PubMed] [Google Scholar]
- Ditta G., Schmidhauser T., Yakobson E., Lu P., Liang X. W., Finlay D. R., Guiney D., Helinski D. R. Plasmids related to the broad host range vector, pRK290, useful for gene cloning and for monitoring gene expression. Plasmid. 1985 Mar;13(2):149–153. doi: 10.1016/0147-619x(85)90068-x. [DOI] [PubMed] [Google Scholar]
- Dross F., Geisler V., Lenger R., Theis F., Krafft T., Fahrenholz F., Kojro E., Duchêne A., Tripier D., Juvenal K. The quinone-reactive Ni/Fe-hydrogenase of Wolinella succinogenes. Eur J Biochem. 1992 May 15;206(1):93–102. doi: 10.1111/j.1432-1033.1992.tb16905.x. [DOI] [PubMed] [Google Scholar]
- Eberz G., Eitinger T., Friedrich B. Genetic determinants of a nickel-specific transport system are part of the plasmid-encoded hydrogenase gene cluster in Alcaligenes eutrophus. J Bacteriol. 1989 Mar;171(3):1340–1345. doi: 10.1128/jb.171.3.1340-1345.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eberz G., Friedrich B. Three trans-acting regulatory functions control hydrogenase synthesis in Alcaligenes eutrophus. J Bacteriol. 1991 Mar;173(6):1845–1854. doi: 10.1128/jb.173.6.1845-1854.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eberz G., Hogrefe C., Kortlüke C., Kamienski A., Friedrich B. Molecular cloning of structural and regulatory hydrogenase (hox) genes of Alcaligenes eutrophus H16. J Bacteriol. 1986 Nov;168(2):636–641. doi: 10.1128/jb.168.2.636-641.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eitinger T., Friedrich B. Cloning, nucleotide sequence, and heterologous expression of a high-affinity nickel transport gene from Alcaligenes eutrophus. J Biol Chem. 1991 Feb 15;266(5):3222–3227. [PubMed] [Google Scholar]
- Ford C. M., Garg N., Garg R. P., Tibelius K. H., Yates M. G., Arp D. J., Seefeldt L. C. The identification, characterization, sequencing and mutagenesis of the genes (hupSL) encoding the small and large subunits of the H2-uptake hydrogenase of Azotobacter chroococcum. Mol Microbiol. 1990 Jun;4(6):999–1008. doi: 10.1111/j.1365-2958.1990.tb00672.x. [DOI] [PubMed] [Google Scholar]
- Friedrich B., Heine E., Finck A., Friedrich C. G. Nickel requirement for active hydrogenase formation in Alcaligenes eutrophus. J Bacteriol. 1981 Mar;145(3):1144–1149. doi: 10.1128/jb.145.3.1144-1149.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedrich C. G. Depression of hydrogenase during limitation of electron donors and derepression of ribulosebisphosphate carboxylase during carbon limitation of Alcaligenes eutrophus. J Bacteriol. 1982 Jan;149(1):203–210. doi: 10.1128/jb.149.1.203-210.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gough J. A., Murray N. E. Sequence diversity among related genes for recognition of specific targets in DNA molecules. J Mol Biol. 1983 May 5;166(1):1–19. doi: 10.1016/s0022-2836(83)80047-3. [DOI] [PubMed] [Google Scholar]
- Henikoff S. Unidirectional digestion with exonuclease III creates targeted breakpoints for DNA sequencing. Gene. 1984 Jun;28(3):351–359. doi: 10.1016/0378-1119(84)90153-7. [DOI] [PubMed] [Google Scholar]
- Hogrefe C., Römermann D., Friedrich B. Alcaligenes eutrophus hydrogenase genes (Hox). J Bacteriol. 1984 Apr;158(1):43–48. doi: 10.1128/jb.158.1.43-48.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hormel S., Walsh K. A., Prickril B. C., Titani K., LeGall J., Sieker L. C. Amino acid sequence of rubredoxin from Desulfovibrio desulfuricans strain 27774. FEBS Lett. 1986 May 26;201(1):147–150. doi: 10.1016/0014-5793(86)80588-9. [DOI] [PubMed] [Google Scholar]
- Knauf V. C., Nester E. W. Wide host range cloning vectors: a cosmid clone bank of an Agrobacterium Ti plasmid. Plasmid. 1982 Jul;8(1):45–54. doi: 10.1016/0147-619x(82)90040-3. [DOI] [PubMed] [Google Scholar]
- Kortlüke C., Friedrich B. Maturation of membrane-bound hydrogenase of Alcaligenes eutrophus H16. J Bacteriol. 1992 Oct;174(19):6290–6293. doi: 10.1128/jb.174.19.6290-6293.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyte J., Doolittle R. F. A simple method for displaying the hydropathic character of a protein. J Mol Biol. 1982 May 5;157(1):105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- Leclerc M., Colbeau A., Cauvin B., Vignais P. M. Cloning and sequencing of the genes encoding the large and the small subunits of the H2 uptake hydrogenase (hup) of Rhodobacter capsulatus. Mol Gen Genet. 1988 Sep;214(1):97–107. doi: 10.1007/BF00340186. [DOI] [PubMed] [Google Scholar]
- Lorenz B., Schneider K., Kratzin H., Schlegel H. G. Immunological comparison of subunits isolated from various hydrogenases of aerobic hydrogen bacteria. Biochim Biophys Acta. 1989 Mar 16;995(1):1–9. doi: 10.1016/0167-4838(89)90225-2. [DOI] [PubMed] [Google Scholar]
- Lutz S., Jacobi A., Schlensog V., Böhm R., Sawers G., Böck A. Molecular characterization of an operon (hyp) necessary for the activity of the three hydrogenase isoenzymes in Escherichia coli. Mol Microbiol. 1991 Jan;5(1):123–135. doi: 10.1111/j.1365-2958.1991.tb01833.x. [DOI] [PubMed] [Google Scholar]
- Mead D. A., Szczesna-Skorupa E., Kemper B. Single-stranded DNA 'blue' T7 promoter plasmids: a versatile tandem promoter system for cloning and protein engineering. Protein Eng. 1986 Oct-Nov;1(1):67–74. doi: 10.1093/protein/1.1.67. [DOI] [PubMed] [Google Scholar]
- Menon A. L., Stults L. W., Robson R. L., Mortenson L. E. Cloning, sequencing and characterization of the [NiFe]hydrogenase-encoding structural genes (hoxK and hoxG) from Azotobacter vinelandii. Gene. 1990 Nov 30;96(1):67–74. doi: 10.1016/0378-1119(90)90342-o. [DOI] [PubMed] [Google Scholar]
- Menon N. K., Robbins J., Peck H. D., Jr, Chatelus C. Y., Choi E. S., Przybyla A. E. Cloning and sequencing of a putative Escherichia coli [NiFe] hydrogenase-1 operon containing six open reading frames. J Bacteriol. 1990 Apr;172(4):1969–1977. doi: 10.1128/jb.172.4.1969-1977.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menon N. K., Robbins J., Wendt J. C., Shanmugam K. T., Przybyla A. E. Mutational analysis and characterization of the Escherichia coli hya operon, which encodes [NiFe] hydrogenase 1. J Bacteriol. 1991 Aug;173(15):4851–4861. doi: 10.1128/jb.173.15.4851-4861.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer J., Gagnon J., Sieker L. C., Van Dorsselaer A., Moulis J. M. Rubredoxin from Clostridium thermosaccharolyticum. Amino acid sequence, mass-spectrometric and preliminary crystallographic data. Biochem J. 1990 Nov 1;271(3):839–841. doi: 10.1042/bj2710839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Normark S., Bergström S., Edlund T., Grundström T., Jaurin B., Lindberg F. P., Olsson O. Overlapping genes. Annu Rev Genet. 1983;17:499–525. doi: 10.1146/annurev.ge.17.120183.002435. [DOI] [PubMed] [Google Scholar]
- Przybyla A. E., Robbins J., Menon N., Peck H. D., Jr Structure-function relationships among the nickel-containing hydrogenases. FEMS Microbiol Rev. 1992 Feb;8(2):109–135. doi: 10.1111/j.1574-6968.1992.tb04960.x. [DOI] [PubMed] [Google Scholar]
- Roth J., Bendayan M., Carlemalm E., Villiger W., Garavito M. Enhancement of structural preservation and immunocytochemical staining in low temperature embedded pancreatic tissue. J Histochem Cytochem. 1981 May;29(5):663–671. doi: 10.1177/29.5.6166664. [DOI] [PubMed] [Google Scholar]
- Russel M., Model P. Replacement of the fip gene of Escherichia coli by an inactive gene cloned on a plasmid. J Bacteriol. 1984 Sep;159(3):1034–1039. doi: 10.1128/jb.159.3.1034-1039.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SCHLEGEL H. G., KALTWASSER H., GOTTSCHALK G. [A submersion method for culture of hydrogen-oxidizing bacteria: growth physiological studies]. Arch Mikrobiol. 1961;38:209–222. [PubMed] [Google Scholar]
- Saeki K., Yao Y., Wakabayashi S., Shen G. J., Zeikus J. G., Matsubara H. Ferredoxin and rubredoxin from Butyribacterium methylotrophicum: complete primary structures and construction of phylogenetic trees. J Biochem. 1989 Oct;106(4):656–662. doi: 10.1093/oxfordjournals.jbchem.a122912. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sayavedra-Soto L. A., Powell G. K., Evans H. J., Morris R. O. Nucleotide sequence of the genetic loci encoding subunits of Bradyrhizobium japonicum uptake hydrogenase. Proc Natl Acad Sci U S A. 1988 Nov;85(22):8395–8399. doi: 10.1073/pnas.85.22.8395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schink B., Schlegel H. G. The membrane-bound hydrogenase of Alcaligenes eutrophus. I. Solubilization, purification, and biochemical properties. Biochim Biophys Acta. 1979 Apr 12;567(2):315–324. doi: 10.1016/0005-2744(79)90117-7. [DOI] [PubMed] [Google Scholar]
- Seki Y., Seki S., Satoh M., Ikeda A., Ishimoto M. Rubredoxin from Clostridium perfringens: complete amino acid sequence and participation in nitrate reduction. J Biochem. 1989 Aug;106(2):336–341. doi: 10.1093/oxfordjournals.jbchem.a122854. [DOI] [PubMed] [Google Scholar]
- Shimizu F., Ogata M., Yagi T., Wakabayashi S., Matsubara H. Amino acid sequence and function of rubredoxin from Desulfovibrio vulgaris Miyazaki. Biochimie. 1989 Nov-Dec;71(11-12):1171–1177. doi: 10.1016/0300-9084(89)90020-5. [DOI] [PubMed] [Google Scholar]
- Simon R., Quandt J., Klipp W. New derivatives of transposon Tn5 suitable for mobilization of replicons, generation of operon fusions and induction of genes in gram-negative bacteria. Gene. 1989 Aug 1;80(1):161–169. doi: 10.1016/0378-1119(89)90262-x. [DOI] [PubMed] [Google Scholar]
- Tabor S., Richardson C. C. A bacteriophage T7 RNA polymerase/promoter system for controlled exclusive expression of specific genes. Proc Natl Acad Sci U S A. 1985 Feb;82(4):1074–1078. doi: 10.1073/pnas.82.4.1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thöny B., Hennecke H. The -24/-12 promoter comes of age. FEMS Microbiol Rev. 1989 Dec;5(4):341–357. doi: 10.1016/0168-6445(89)90028-4. [DOI] [PubMed] [Google Scholar]
- Tran-Betcke A., Warnecke U., Böcker C., Zaborosch C., Friedrich B. Cloning and nucleotide sequences of the genes for the subunits of NAD-reducing hydrogenase of Alcaligenes eutrophus H16. J Bacteriol. 1990 Jun;172(6):2920–2929. doi: 10.1128/jb.172.6.2920-2929.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woolley K. J., Meyer T. E. The complete amino acid sequence of rubredoxin from the green phototrophic bacterium Chlorobium thiosulphatophilum strain PM. Eur J Biochem. 1987 Feb 16;163(1):161–166. doi: 10.1111/j.1432-1033.1987.tb10750.x. [DOI] [PubMed] [Google Scholar]