Abstract
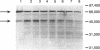
Pyrodictium brockii is a hyperthermophilic archaebacterium with an optimal growth temperature of 105 degrees C. P. brockii is also a chemolithotroph, requiring H2 and CO2 for growth. We have purified the hydrogen uptake hydrogenase from membranes of P. brockii by reactive red affinity chromatography and sucrose gradient centrifugation. The molecular mass of the holoenzyme was 118,000 +/- 19,000 Da in sucrose gradients. The holoenzyme consisted of two subunits by sodium dodecyl sulfate-polyacrylamide gel electrophoresis. The large subunit had a molecular mass of 66,000 Da, and the small subunit had a molecular mass of 45,000 Da. Colorometric analysis of Fe and S content in reactive red-purified hydrogenase revealed 8.7 +/- 0.6 mol of Fe and 6.2 +/- 1.2 mol of S per mol of hydrogenase. Growth of cells in 63NiCl2 resulted in label incorporation into reactive red-purified hydrogenase. Growth of cells in 63NiCl2 resulted in label incorporation into reactive red-purified hydrogenase. Temperature stability studies indicated that the membrane-bound form of the enzyme was more stable than the solubilized purified form over a period of minutes with respect to temperature. However, the membranes were not able to protect the enzyme from thermal inactivation over a period of hours. The artificial electron acceptor specificity of the pure enzyme was similar to that of the membrane-bound form, but the purified enzyme was able to evolve H2 in the presence of reduced methyl viologen. The Km of membrane-bound hydrogenase for H2 was approximately 19 microM with methylene blue as the electron acceptor, whereas the purified enzyme had a higher Km value.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Adams M. W., Mortenson L. E., Chen J. S. Hydrogenase. Biochim Biophys Acta. 1980 Dec;594(2-3):105–176. doi: 10.1016/0304-4173(80)90007-5. [DOI] [PubMed] [Google Scholar]
- Alex L. A., Reeve J. N., Orme-Johnson W. H., Walsh C. T. Cloning, sequence determination, and expression of the genes encoding the subunits of the nickel-containing 8-hydroxy-5-deazaflavin reducing hydrogenase from Methanobacterium thermoautotrophicum delta H. Biochemistry. 1990 Aug 7;29(31):7237–7244. doi: 10.1021/bi00483a011. [DOI] [PubMed] [Google Scholar]
- Aono S., Bryant F. O., Adams M. W. A novel and remarkably thermostable ferredoxin from the hyperthermophilic archaebacterium Pyrococcus furiosus. J Bacteriol. 1989 Jun;171(6):3433–3439. doi: 10.1128/jb.171.6.3433-3439.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arp D. J., Burris R. H. Kinetic mechanism of the hydrogen-oxidizing hydrogenase from soybean nodule bacteroids. Biochemistry. 1981 Apr 14;20(8):2234–2240. doi: 10.1021/bi00511a025. [DOI] [PubMed] [Google Scholar]
- Arp D. J., Burris R. H. Purification and properties of the particulate hydrogenase from the bacteroids of soybean root nodules. Biochim Biophys Acta. 1979 Oct 11;570(2):221–230. doi: 10.1016/0005-2744(79)90142-6. [DOI] [PubMed] [Google Scholar]
- Baron S. F., Ferry J. G. Purification and properties of the membrane-associated coenzyme F420-reducing hydrogenase from Methanobacterium formicicum. J Bacteriol. 1989 Jul;171(7):3846–3853. doi: 10.1128/jb.171.7.3846-3853.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beinert H. Semi-micro methods for analysis of labile sulfide and of labile sulfide plus sulfane sulfur in unusually stable iron-sulfur proteins. Anal Biochem. 1983 Jun;131(2):373–378. doi: 10.1016/0003-2697(83)90186-0. [DOI] [PubMed] [Google Scholar]
- Blumentals I. I., Robinson A. S., Kelly R. M. Characterization of sodium dodecyl sulfate-resistant proteolytic activity in the hyperthermophilic archaebacterium Pyrococcus furiosus. Appl Environ Microbiol. 1990 Jul;56(7):1992–1998. doi: 10.1128/aem.56.7.1992-1998.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown S. H., Kelly R. M. Cultivation Techniques for Hyperthermophilic Archaebacteria: Continuous Culture of Pyrococcus furiosus at Temperatures near 100 degrees C. Appl Environ Microbiol. 1989 Aug;55(8):2086–2088. doi: 10.1128/aem.55.8.2086-2088.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryant F. O., Adams M. W. Characterization of hydrogenase from the hyperthermophilic archaebacterium, Pyrococcus furiosus. J Biol Chem. 1989 Mar 25;264(9):5070–5079. [PubMed] [Google Scholar]
- Chen J. S., Mortenson L. E. Inhibition of methylene blue formation during determination of the acid-labile sulfide of iron-sulfur protein samples containing dithionite. Anal Biochem. 1977 May 1;79(1-2):157–165. doi: 10.1016/0003-2697(77)90390-6. [DOI] [PubMed] [Google Scholar]
- Conover R. C., Kowal A. T., Fu W. G., Park J. B., Aono S., Adams M. W., Johnson M. K. Spectroscopic characterization of the novel iron-sulfur cluster in Pyrococcus furiosus ferredoxin. J Biol Chem. 1990 May 25;265(15):8533–8541. [PubMed] [Google Scholar]
- Costantino H. R., Brown S. H., Kelly R. M. Purification and characterization of an alpha-glucosidase from a hyperthermophilic archaebacterium, Pyrococcus furiosus, exhibiting a temperature optimum of 105 to 115 degrees C. J Bacteriol. 1990 Jul;172(7):3654–3660. doi: 10.1128/jb.172.7.3654-3660.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiebig K., Friedrich B. Purification of the F420-reducing hydrogenase from Methanosarcina barkeri (strain Fusaro). Eur J Biochem. 1989 Sep 1;184(1):79–88. doi: 10.1111/j.1432-1033.1989.tb14992.x. [DOI] [PubMed] [Google Scholar]
- Fox J. A., Livingston D. J., Orme-Johnson W. H., Walsh C. T. 8-Hydroxy-5-deazaflavin-reducing hydrogenase from Methanobacterium thermoautotrophicum: 1. Purification and characterization. Biochemistry. 1987 Jul 14;26(14):4219–4227. doi: 10.1021/bi00388a007. [DOI] [PubMed] [Google Scholar]
- Hausinger R. P. Nickel utilization by microorganisms. Microbiol Rev. 1987 Mar;51(1):22–42. doi: 10.1128/mr.51.1.22-42.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LOVENBERG W., BUCHANAN B. B., RABINOWITZ J. C. STUDIES ON THE CHEMICAL NATURE OF CLOSTRIDIAL FERREDOXIN. J Biol Chem. 1963 Dec;238:3899–3913. [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- MARTIN R. G., AMES B. N. A method for determining the sedimentation behavior of enzymes: application to protein mixtures. J Biol Chem. 1961 May;236:1372–1379. [PubMed] [Google Scholar]
- Maier R. J., Merberg D. M. Rhizobium japonicum mutants that are hypersensitive to repression of H2 uptake by oxygen. J Bacteriol. 1982 Apr;150(1):161–167. doi: 10.1128/jb.150.1.161-167.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Brian M. R., Maier R. J. Hydrogen metabolism in Rhizobium: energetics, regulation, enzymology and genetics. Adv Microb Physiol. 1988;29:1–52. doi: 10.1016/s0065-2911(08)60345-8. [DOI] [PubMed] [Google Scholar]
- Pihl T. D., Schicho R. N., Kelly R. M., Maier R. J. Characterization of hydrogen-uptake activity in the hyperthermophile Pyrodictium brockii. Proc Natl Acad Sci U S A. 1989 Jan;86(1):138–141. doi: 10.1073/pnas.86.1.138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polacheck I., Cabib E. A simple procedure for protein determination by the Lowry method in dilute solutions and in the presence of interfering substances. Anal Biochem. 1981 Nov 1;117(2):311–314. doi: 10.1016/0003-2697(81)90784-3. [DOI] [PubMed] [Google Scholar]
- Reeve J. N., Beckler G. S., Cram D. S., Hamilton P. T., Brown J. W., Krzycki J. A., Kolodziej A. F., Alex L., Orme-Johnson W. H., Walsh C. T. A hydrogenase-linked gene in Methanobacterium thermoautotrophicum strain delta H encodes a polyferredoxin. Proc Natl Acad Sci U S A. 1989 May;86(9):3031–3035. doi: 10.1073/pnas.86.9.3031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schink B., Schlegel H. G. The membrane-bound hydrogenase of Alcaligenes eutrophus. I. Solubilization, purification, and biochemical properties. Biochim Biophys Acta. 1979 Apr 12;567(2):315–324. doi: 10.1016/0005-2744(79)90117-7. [DOI] [PubMed] [Google Scholar]
- Schneider K., Pinkwart M., Jochim K. Purification of hydrogenases by affinity chromatography on Procion Red-agarose. Biochem J. 1983 Aug 1;213(2):391–398. doi: 10.1042/bj2130391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah N. N., Clark D. S. Partial Purification and Characterization of Two Hydrogenases from the Extreme Thermophile Methanococcus jannaschii. Appl Environ Microbiol. 1990 Apr;56(4):858–863. doi: 10.1128/aem.56.4.858-863.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sim E., Vignais P. M. Comparison of the membrane-bound and detergent-solubilised hydrogenase from paracoccus denitrificans. Isolation of the hydrogenase. Biochim Biophys Acta. 1979 Sep 12;570(1):43–55. doi: 10.1016/0005-2744(79)90199-2. [DOI] [PubMed] [Google Scholar]
- Steigerwald V. J., Beckler G. S., Reeve J. N. Conservation of hydrogenase and polyferredoxin structures in the hyperthermophilic archaebacterium Methanothermus fervidus. J Bacteriol. 1990 Aug;172(8):4715–4718. doi: 10.1128/jb.172.8.4715-4718.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stults L. W., Moshiri F., Maier R. J. Aerobic purification of hydrogenase from Rhizobium japonicum by affinity chromatography. J Bacteriol. 1986 Jun;166(3):795–800. doi: 10.1128/jb.166.3.795-800.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stults L. W., O'Hara E. B., Maier R. J. Nickel is a component of hydrogenase in Rhizobium japonicum. J Bacteriol. 1984 Jul;159(1):153–158. doi: 10.1128/jb.159.1.153-158.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woese C. R. Bacterial evolution. Microbiol Rev. 1987 Jun;51(2):221–271. doi: 10.1128/mr.51.2.221-271.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]