Abstract
Previous findings have suggested that class IIa histone deacetylases (HDACs) (HDAC4, -5, -7, and -9) are inactive on acetylated substrates, thus differing from class I and IIb enzymes. Here, we present evidence supporting this view and demonstrate that class IIa HDACs are very inefficient enzymes on standard substrates. We identified HDAC inhibitors unable to bind recombinant human HDAC4 while showing inhibition in a typical HDAC4 enzymatic assay, suggesting that the observed activity rather reflects the involvement of endogenous copurified class I HDACs. Moreover, an HDAC4 catalytic domain purified from bacteria was 1,000-fold less active than class I HDACs on standard substrates. A catalytic Tyr is conserved in all HDACs except for vertebrate class IIa enzymes where it is replaced by His. Given the high structural conservation of HDAC active sites, we predicted the class IIa His-Nε2 to be too far away to functionally substitute the class I Tyr-OH in catalysis. Consistently, a Tyr-to-His mutation in class I HDACs severely reduced their activity. More importantly, a His-976-Tyr mutation in HDAC4 produced an enzyme with a catalytic efficiency 1,000-fold higher than WT, and this “gain of function phenotype” could be extended to HDAC5 and -7. We also identified trifluoroacetyl-lysine as a class IIa-specific substrate in vitro. Hence, vertebrate class IIa HDACs may have evolved to maintain low basal activities on acetyl-lysines and to efficiently process restricted sets of specific, still undiscovered natural substrates.
Keywords: catalytic domain, enzymatic activity, trifluoroacetyl-lysine, gain of function
Lysine acetylation, the transfer of an acetyl moiety from acetyl-CoA to the ε-amino group of specific lysine residues, has recently emerged as a major form of posttranslational modification regulating functions of core histones and an increasing number of transcription factors and other cellular and viral proteins (1). Acetylation, catalyzed by histone acetyl transferases, is a dynamic process reversed by a class of enzymes known as histone deacetylases (HDACs) (2, 3).
Enzymes of the HDAC superfamily can be divided into two large groups (classes I and II) based on characteristic, conserved sequence motifs within a domain of ≈350 aa harbouring the known or putative catalytic HDAC domain (3, 4). A subset of class I members containing HDAC11-related enzymes has recently been proposed as an independent group (class IV) (5). All three classes are distinct from the sirtuin-family enzymes (class III) (6) in the catalytic domain primary sequence and three-dimensional structure, and in the catalytic mechanism. Several HDAC structures confirm the presence of a common catalytic site where a Zn ion coordinated by highly conserved residues contacts the hydroxamate moiety of HDAC inhibitors (HDACis) (7–10). On this basis, a common enzymatic mechanism has been proposed focused on the Zn-catalyzed hydrolysis of the acetyl-lysine amide bond (7).
Metazoan class II HDACs are further divided into subclasses IIa and IIb. Class IIa members (HDAC4, -5, -7, and -9 in mammals) form a distinct subgroup because of the presence, in addition to the HDAC domain, of an extended N-terminal regulatory domain. Although the role of class IIa HDACs in tissue-specific gene regulation is well documented (11–14), the contribution of the HDAC domain to this activity is still controversial. There is ample evidence that class IIa members exert transcriptional repression because of their ability to directly inactivate specific transcription factors. In addition, they can recruit a number of distinct corepressors and/or protein-modifying enzymes that, in turn, inactivate target transcription factors (4, 15). Most of these protein–protein interactions occur via the N-terminal domain, whereas the presence of the catalytic domain is often not strictly required. A natural HDAC9 splice variant lacking the HDAC domain [myocyte enhancer factor-2 interacting transcription repressor/HDAC-related protein (MITR/HDRP)] as well as analogous artificially truncated forms of other class IIa members retain full transcriptional repressive functions (16, 17). In addition, several reports have questioned whether class IIa HDACs possess an intrinsic deacetylase activity, proposing instead that they function through additional associated HDACs. All class IIa members expressed in mammalian cells are known to recruit class I HDACs, which possess high deacetylase activities both in vitro and in cells (18–20). In particular, HDAC4 and -7 were shown to associate with HDAC3–SMRT/N–CoR complexes through their HDAC domains (19, 20). Site-directed mutagenesis of conserved active site residues in HDAC4 failed to clarify this point in that all HDAC4 mutants showing impaired deacetylase activity in vitro also lost the ability to interact with HDAC3–SMRT/N–CoR complexes (20). Consequently, it remains unclear whether the resulting drop in overall deacetylase activity was caused by a loss of intrinsic HDAC4 activity or loss of the associated highly active HDAC3 complex. Attempts to obtain active recombinant class IIa HDACs in heterologous systems were also unsuccessful (21). Whether this is due to a lack of putative cofactors required to boost class IIa HDAC intrinsic activity or a lack of endogenous class I enzymes is still unknown. An additional limitation is the lack of isotype/class-specific HDACis that may help distinguishing the contribution of class IIa intrinsic catalytic activity from that of associated class I HDACs.
We addressed these issues by using HDAC4 as a representative class IIa model system with four different approaches: (i) comparison of the inhibitory potential of selected HDACis on HDAC4-associated deacetylase activity with their capability to bind HDAC4 catalytic domain; (ii) functional analysis of a purified HDAC4 catalytic domain expressed in Escherichia coli; (iii) structure-guided mutagenesis of a single catalytic residue present only in vertebrate class IIa HDACs; and (iv) screening for HDAC4-specific substrates. Collectively, our findings, extended also to other class IIa enzymes, led to the following conclusions. Class IIa HDACs possess a weak but measurable intrinsic capability of hydrolyzing acetyl-lysines in vitro. Most of the enzymatic activity associated with class IIa HDACs ectopically expressed in mammalian cells is due to endogenous class I HDACs present in class IIa immunocomplexes. More importantly, we could completely ascribe the low catalytic efficiency of mammalian class IIa enzymes to the presence of a unique H residue replacing the catalytic Y conserved in all other prokaryotic and eukaryotic deacetylases. Indeed, catalytic activity toward canonical HDAC substrates could be restored by a single H-to-Y substitution in the active site. Finally, we have identified trifluoroacetyl-lysine as a class IIa-specific substrate in vitro.
Results
Sequence and Structure Analysis Indicates that Vertebrate Class IIa HDACs Have Distinct Properties.
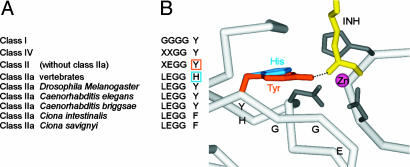
Despite the differences in sequence motifs distinguishing class I and II HDACs [supporting information (SI) Fig. 7], the active site is structurally highly conserved, and all functional residues are in equivalent positions. A common feature of all HDAC structures is the presence of a Y residue postulated to act as a transition-state stabilizer (7). However, examination of sequence conservation across many HDACs revealed an unexpected feature exclusive to vertebrate class IIa HDACs (Fig. 1A). In the vertebrate class IIa HDACs, the catalytic Y is replaced by an H, a conservative substitution, whereas other functional residues are virtually always conserved (SI Fig. 7). In the structures of the bacterial HDAC homolog HDLP (7), HDAC8 (9, 10), and a class II bacterial homolog (8), this Y is located close to the end of a β-strand, a structural element conserved among all these homologs including the local backbone conformation (data not shown). To act as transition-state stabilizer, the Y side chain has to be oriented along the β-strand, as seen in all these crystal structures. For vertebrate class IIa HDACs, the Y is replaced by an H side chain, which is too short to reach into the active site (Fig. 1B). In class IIa HDACs, three different residues (Y, H, and F) are observed at this position and, given the particular structural context, it is unlikely that for each residue a concomitant conformational change would occur.
Fig. 1.
Sequence motif conservation and active-site geometry. (A) Class-specific sequence motif completely conserved in class I. Conservation is generally high also within class II, with the notable exception of class IIa. (B) Active-site region of a bacterial class II homolog. To substitute Y as transition-state stabilizer, the H has to move closer to the active site. The LEGGY motif is indicated, as is the interaction between the Y and a representative hydroxamic acid moiety (INH).
Consequently, we hypothesized that the catalytic domain of vertebrate class IIa HDACs should be less effective in processing canonical HDAC substrates containing acetyl-lysines. Nevertheless, the high conservation of active-site and substrate-binding residues (SI Fig. 7) suggests that class IIa HDACs should be functional enzymes.
Class I HDACs with the Y Mutated into H Are Inactive on Canonical HDAC Substrates.
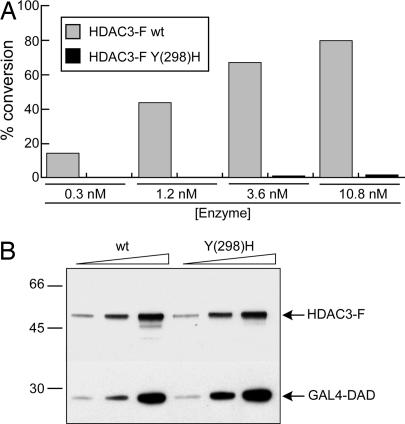
One implication of our hypothesis was that class I enzymatic activities should be affected by substituting the catalytic Y with an H. We therefore mutated Y-298 into H in the active site of HDAC3, a representative class I member. WT and mutant flag-tagged HDAC3 enzymes were coexpressed in HeLa cells and coimmunopurified together with a GAL4 DBD fusion of the N-CoR Deacetylase Activation Domain (N-CoR DAD) (22, 23). As predicted, replacing the active site Y with H was sufficient to abolish the enzymatic activity on a histone H4-derived peptide containing a single acetyl-lysine (pepH4-AcK16) (Fig. 2A). Identical results were obtained on3[H]-acetylated core histones (data not shown). Mutations of HDAC active-site residues are reported to impair interactions with protein-binding partners, suggesting that they may also cause alterations in the overall catalytic domain structure (24). However, the Y298H substitution in HDAC3 did not affect its association with N-CoR DAD (Fig. 2B), suggesting that the integrity of the catalytic domain was preserved. Furthermore, replacement of Y with H produced a loss-of-function phenotype as severe as that caused by replacement with F (SI Fig. 8A). Although similar in size and shape, F lacks the functional hydroxyl group present in Y and therefore cannot contribute to catalysis as a transition-state stabilizer. This mutant also preserved the ability to bind N-CoR DAD (SI Fig. 8B), thus confirming that the substitution with F is conservative regarding the active site geometry. Finally, we demonstrated a more general role for the catalytic Y by mutating Y-303 into H in the active site of HDAC1-FLAG. As shown in SI Fig. 8C, the mutation actually reduced HDAC1 deacetylase activity 5- to 8-fold. Notably, key interactions with different endogenous HDAC1/2 binding proteins and with HDAC2 were not affected by the mutation (SI Fig. 8D). Indeed, the residual activity shown by the mutant can probably be ascribed to associated native HDAC2.
Fig. 2.
The HDAC3 Y(298)H substitution generates a loss-of-function mutant. (A) Enzymatic activities of WT and Y(298)H-mutated HDAC3-FLAG (HDAC3-F) proteins on pepH4-AcK16. (B) Western blot of HDAC3-FLAG immunocomplexes assayed in A by using anti-FLAG (HDAC3-F) or anti-Gal4 (GAL4-DAD) antibodies. Ten, 20, and 40 nM WT and mutant HDAC3 were analyzed.
Class IIa HDACs with the H Mutated into F Are Not Impaired in Their Catalytic Activity on Canonical HDAC Substrates.
Having demonstrated that Y mutants of class I HDACs lose most of their canonical catalytic activity, we used a similar approach to study the intrinsic enzymatic activity of HDAC4. Ectopically expressed class IIa enzymes are generally found to be associated with highly active endogenous class I enzymes such as HDAC1, -2, and -3 (refs. 18–20 and data not shown). Measuring the deacetylase activity of HDAC4 complexes purified from mammalian cells would therefore only reflect the overall activity of the complex and not necessarily that of HDAC4. On the other hand, if HDAC4 contributes significantly to this activity, a mutation of H into F, analogous to class I mutants, should have a strong impact on the overall deacetylase activity. C-terminally flag-tagged HDAC4 proteins, either WT or H976F, were purified from HEK293 cells. As shown in SI Fig. 8E, mutation into F did not alter the activity of the purified HDAC4 complex on pepH4-AcK16. Interestingly, the F mutant preserved WT interactions with the endogenous HDAC3–N–CoR complex (SI Fig. 8F) that, according to published data, should be mainly responsible for the activity on acetylated substrates (20).
Inhibition by Selected HDACis of HDAC4-Associated Activity Does Not Correlate with Their Binding to the HDAC4 Catalytic Domain.
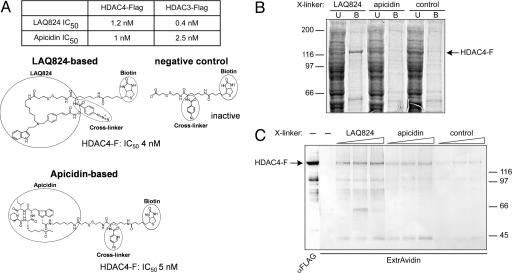
Independent evidence confirmed that HDAC4-FLAG does not significantly contribute to the observed enzymatic activity, which rather reflects the involvement of endogenous copurified class I HDACs. Two structurally dissimilar HDACis, the hydroxamate LAQ824 and the cyclic tetrapeptide apicidin, were selected as test molecules for binding studies. Both compounds showed similar nanomolar potencies in inhibiting the deacetylase activity of ectopically expressed HDAC3-FLAG and HDAC4-FLAG (Fig. 3A). Therefore, they were modified to include a photoactivatable cross-linker and the trapping molecule biotin (Fig. 3A). An identical modified molecule lacking the HDACi moiety was included as negative control. Direct binding to HDAC4-FLAG was measured in two ways: (i) in pull-down experiments by capturing HDAC4-FLAG from a cell extract on streptavidin-coated beads preadsorbed with the biotinylated derivatives (Fig. 3B); and (ii) by UV cross-linking and analysis of the protein covalently bound to the biotinylated compounds in ExtrAvidin-Western blots (Fig. 3C). Although LAQ824 and apicidin, as well as their derivatives, were equipotent in inhibiting HDAC4-FLAG-associated activity (Fig. 3A), of the two, only the LAQ824 derivative significantly and specifically bound HDAC4-FLAG (Fig. 3 B and C). UV cross-linking experiments with HDAC3-FLAG and HDAC6-FLAG showed instead a perfect correspondence between sensitivity and binding to both compounds (SI Fig. 9A). These findings were confirmed and extended by competition experiments in which binding to the LAQ824-based compound was challenged by adding different HDACis at suitable molar excesses (SI Fig. 9 B and C). Table 1 summarizes comparative data of the binding and inhibition properties of selected compounds on HDAC4-FLAG. The lack of correlation between binding and inhibition observed with apicidins and MS-275 confirmed that HDAC4 does not contribute to the deacetylase activity of the HDAC4 complex (or does so only to a marginal extent).
Fig. 3.
HDAC4 compounds binding assays. (A) IC50 values of LAQ824 and apicidin on HDAC4-FLAG and HDAC3-FLAG activities and structures of the corresponding linkerized/biotinylated derivatives. A negative control lacking the HDACi moiety is also shown. The IC50 values of the modified compounds for HDAC4-FLAG (HDAC4-F) are reported. (B) HDAC4-FLAG pull-down on streptavidin-coated magnetic beads preabsorbed with each of the three biotinylated compounds shown in A. A representative Coomassie blue-stained SDS/PAGE gel loaded with the unbound (U) and bound (B) fractions is shown. (C) UV cross-linking of HDAC4-FLAG to each of the three compounds described in A. A representative ExtrAvidin Western blot is shown. A control lane developed with anti-FLAG antibodies after a mock UV treatment is also shown.
Table 1.
Binding and inhibitory activities of selected HDACi on different HDAC4 variants
| Compound | Binding assay HDAC4-F + CD | Histone assay HDAC4-F* | Biomol assay HDAC4-F* | Biomol assay mHDAC4-F† | Biomol assay mHDAC4-CD† |
|---|---|---|---|---|---|
| MS-275 | − | 510 (±62) | 3,017 (±422) | NA up to 10,000 | NA up to 10,000 |
| Apicidin | − | 2.4 (±0.5) | 1.2 (±0.3) | NA up to 10,000 | NA up to 10,000 |
| Open Apicidin | − | 152 (±24) | 88 (±11) | NA up to10,000 | NA up to 10,000 |
| LAQ824 | + | 4 (±1.3) | 1 (±0.5) | 3 (±0.8) | 2.5 (±0.7) |
IC50 values (nM) are the means of at least two independent experiments. NA, not active.
*[HDAC4-F] was 15 nM; in these assays endogenous class I HDAC activity is measured.
†[mHDAC4] was 1 nM.
HDAC4 Exhibits Weak Enzymatic Activity on Canonical HDAC Substrates in the Absence of Associated Endogenous Class I HDACs.
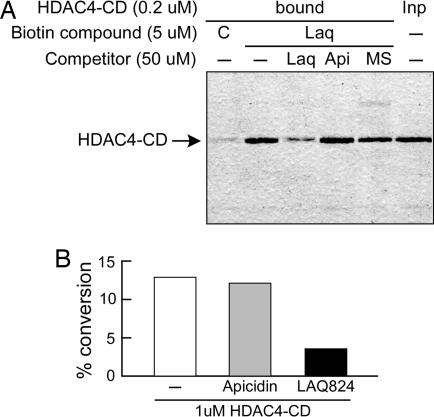
A third and even stronger evidence for WT HDAC4 lacking canonical deacetylase activity came from experiments on the isolated catalytic domain. Based on HDAC sequence homologies, an N-terminally truncated HDAC4 catalytic domain (HDAC4-CD) starting at Thr 653 was cloned, expressed in E. coli, and purified to homogeneity. This protein was efficiently pulled-down on biotinylated LAQ824 beads (Fig. 4A), suggesting that the structural integrity of the catalytic domain was preserved in the recombinant truncated mutant. Pull-down was significantly reduced by preincubation with an excess of free LAQ824, whereas apicidin and MS275 did not show competition, thus confirming and directly validating the results obtained with ectopic HDAC4-FLAG. When challenged in activity assays, HDAC4-CD showed a measurable deacetylase activity both in the histone-based (Fig. 4B) and the commercial (SI Fig. 10A) HDAC assays. However, this activity was at least 1,000-fold lower than that of class I HDACs ectopically expressed in mammalian cells (SI Fig. 10B) and 100-fold lower than that associated with the HDAC4-FLAG complex purified from mammalian cells (data not shown). In addition, HDAC4-CD was completely inactive on pepH4-AcK16 and on a number of acetylated peptides grafted from nonhistonic HDAC substrates (data not shown). Interestingly, apicidin, a potent inhibitor of the HDAC4-FLAG complex activity (Fig. 3A), was instead unable to affect the enzymatic activity of the isolated domain (Fig. 4B and SI Fig. 10A), consistent with the observed lack of binding.
Fig. 4.
Functional characterization of the isolated HDAC4-CD purified from E. coli. (A) Pull-down of HDAC4-CD on biotinylated LAQ824-coated beads after preincubation with the indicated HDACis [LAQ824 (Laq), apicidin (Api), or MS275 (MS)]. In lane C, control beads coated with the compound lacking the HDACi moiety were used. A representative Coomassie blue-stained SDS/PAGE gel is shown. (B) Enzymatic activity of HDAC4-CD on acetylated histones. One micromolar protein was needed to measure significant activity levels. Deacetylation was assayed in the absence or presence of 10 μM apicidin (inactive) or LAQ824 (active).
Together, these results demonstrate that HDAC4 intrinsic catalytic activity is low on canonical acetylated substrates and does not contribute significantly to the overall activity observed in purified ectopic HDAC4 complexes.
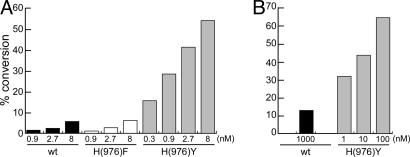
An HDAC4 H976Y Mutation Restores Canonical Deacetylase Activity.
According to the structural predictions, the only feature missing in the catalytic domain of HDAC4 for being a bona fide HDAC is the Y residue involved in stabilizing the transition-state intermediate. Consequently, the substitution of H-976 with Y in HDAC4 was expected to represent a “gain-of-function” mutation. H976Y mutants were therefore generated and expressed as HDAC4-FLAG in mammalian cells and HDAC4-CD in E. coli and tested for histone deacetylase activity. As predicted, both mutated enzymes now showed high deacetylase activity, increased >20-fold for HDAC4-FLAG and >1,000-fold for HDAC4-CD in the linear range of the reaction (Fig. 5 A and B, respectively), reaching levels comparable to class I enzymes. This experimental evidence also proved the HDAC4 catalytic domain to be properly folded under our experimental conditions, because a single amino acid substitution was sufficient to confer high enzymatic activity. Y-substituted HDAC4 proteins maintained their gain-of-function phenotype also on the Fluor de Lys substrate, allowing comparative measurements of inhibition constants for a panel of well known HDACis (Table 1, mHDAC4). Different than WT HDAC4-FLAG, Y mutants were sensitive only to the inhibitory effect of LAQ824, but they were highly resistant to inhibition by compounds belonging to different chemical classes (MS-275, apicidin, and its open derivative), all of which are active on class I HDACs (data not shown). Remarkably, compounds that did not inhibit the H976Y-substituted enzymes were those that did not bind WT HDAC4, thus implying that the mutant proteins retain the WT selectivity of interactions. Furthermore, the HDAC4-CD mutant showed an inhibition profile identical to that of the full-length HDAC4-FLAG mutant, suggesting that the observed selectivity is directly related to the catalytic domain and does not depend on additional N-terminal regions.
Fig. 5.
The HDAC4 H976Y substitution generates a gain-of-function mutant. (A) Comparison of the enzymatic activities exhibited by WT, H976F-mutated, and H976Y-mutated HDAC4-FLAG proteins immunopurified from mammalian cells. (B) Comparison of the enzymatic activities of WT and H976Y-mutated HDAC4-CD proteins purified from E. coli. Data obtained on acetylated histones are shown.
To assess whether this gain-of-function effect is a general property of class IIa enzymes, we performed equivalent experiments on HDAC5 H1006Y and HDAC7 H843Y purified from mammalian cells (SI Fig. 11 A and B, respectively). Again Y mutants exhibited strongly increased activity compared with WT and, like the HDAC4 mutant, could be inhibited by LAQ824 but were resistant to apicidin.
HDAC4-Mediated Transcriptional Repression Does Not Depend on Its Catalytic Activity.
It is well documented that class IIa HDACs are potent and selective transcriptional repressors (4). Having ascertained that HDAC4 is not a proficient deacetylase on canonical substrates in vitro, we asked to what extent its enzymatic activity is responsible for the transcriptional repression exerted in HeLa cells expressing a MEF2-dependent reporter gene (SI Fig. 12). The H976Y mutation did not enhance HDAC4 transcriptional inhibitory activity, despite its clear effect on the enzymatic activity in vitro (SI Fig. 12A). In addition, treatment with HDACis partially relieved the observed repression, implying the involvement of an active deacetylase (SI Fig. 12B). However, LAQ824 and apicidin had similar effects in this assay in contrast to the different inhibitory potencies and binding affinities shown in vitro. These findings support consolidated evidence that the catalytic activity of HDAC4 is dispensable for transcriptional repression and suggest instead a role for one or more class I enzymes present in the HDAC4 multiprotein complex.
Identification of a Class IIa-Specific Substrate.
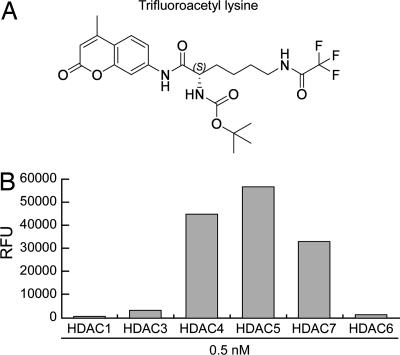
Having ascertained that class IIa HDACs are not proficient enzymes on acetyl-lysines, we sought alternative in vitro substrates. By screening a panel of ≈20 acylated lysine-like molecules, we identified only one substrate, a trifluoroacetyl-lysine, on which both full-length and the catalytic domain of HDAC4 were highly active (Fig. 6 and data not shown). Moreover, this compound was actively and specifically processed by three different class IIa enzymes, whereas it was a poor substrate for both class I and class IIb HDACs (Fig. 6).
Fig. 6.
Trifluoroacetyl-lysine is a class IIa-specific substrate in vitro. (A) The fluorogenic derivative of the trifluoroacetyl-lysine compound is shown. (B) The catalytic activities of flag-tagged HDACs representative of class I, IIa, and IIb were measured on the trifluoroacetyl-lysine substrate shown in A.
Discussion
The conservation of active-site residues and the concomitant preservation of structural features in the active site is a hallmark of many enzymes. Initially, HDACs seem to follow this trend because crystal structures of their catalytic domains confirm a highly conserved active site and substrate-binding pocket. The conservation extends across classes I, II, and IV despite the presence of some class-specific sequence motifs and the generally low homology between classes. In the HDAC active site, a central motif of completely conserved residues coordinates a Zn ion and is complemented by a second group of highly conserved residues involved in the enzymatic reaction and substrate binding. One of these residues, a Y, had been postulated to act as a transition-state stabilizer because of its position in various enzyme-inhibitor complexes. Although generally highly conserved, this residue is substituted by H in all vertebrate class IIa HDACs. Examination of the available structures showed that H, a conservative substitution of Y, should not be able to act as a transition-state stabilizer, thus compromising enzymatic activity according to the proposed mechanism. The picture that emerged from this analysis for HDAC4 (and other vertebrate class IIa enzymes) is that of a theoretically fully functional enzyme if not for the lack of this single important detail.
This prediction was substantiated experimentally in class I enzymes that lost enzymatic activity when mutating Y into H (or F). Further, our hypothesis also predicted that, by changing the H into Y in vertebrate class IIa enzymes, a completely functional active site should be generated. Indeed, in contrast to their WT counterparts, class IIa HDAC Y mutants became fully proficient enzymes with respect to their Lys deacetylase activity on canonical substrates. This finding was confirmed both in the isolated catalytic domain expressed in a prokaryotic system as well as for the full-length enzymes expressed in mammalian cells, including HDAC5 and -7.
We have identified a class IIa-specific synthetic substrate, trifluoroacetyl-lysine. Mechanistically, the presence of the trifluoro group should destabilize the amide bond and thus favor the reaction even in the absence of transition-state stabilization, as postulated for the His-containing class IIa HDACs. Accordingly, potential class IIa-specific natural substrates mimicking the trifluoroacetyl-lysine might contain a hydrolyzable bond intrinsically less stable than an amide bond.
In addition to their known function as transcriptional repressors acting in cohort with associated class I enzymes and directed toward Lys deacetylation, our results point to a previously unknown, specific catalytic activity of vertebrate class IIa HDACs. Specificity is apparently dictated by a single amino acid change in an otherwise conserved active site. Sequence analysis further suggests this “H-associated” specificity is restricted to vertebrates. Invertebrates like Caenorhabditis elegans and Drosophila melanogaster conserve the canonical Y and, accordingly, should retain canonical HDAC catalytic activity as suggested previously (25, 26). The emergent theme predicts class IIa HDACs with intrinsically different catalytic activities for the two groups: (i) canonical HDAC activity in invertebrates and (ii) a more specialized enzymatic activity in vertebrates. Yet another residue, F, is instead found in class IIa homologs of the ascidians Ciona intestinalis, Ciona savignyi, and Halocynthia roretzi. Based on our results, the presence of an F should be incompatible with activity, provided that the catalytic mechanism is conserved. Interestingly, ascidian class IIa sequences also lack one of the conserved Zn ligands, a D is replaced by an S (SI Fig. 7). The evolutionary conservation of this apparently defective active-site configuration is rather unexpected as are its functional implications.
The identification of additional biological activities/substrates that depend on class IIa H-associated activity in mammalian cells requires further work. Ectopically expressed HDAC4 and -5 have been shown to deacetylate Runx3 (27). However, those experiments did not allow discrimination of class IIa HDAC intrinsic deacetylase activities from those of associated class I endogenous enzymes. During the screening of a library of acetylated peptides representative of the known “acetylome,” we have indeed observed (P.G., A.L., C.P., and C.S., unpublished data) that HDAC3, and not HDAC4, -5, -7, or the HDAC3 Y298F mutant, is able to efficiently deacetylate two of the three Runx3 K residues (K148 and K186) previously shown to be acetylated (27). More recently, it has been shown that both Sirt1 (28) and HDAC3 (29), and not HDAC4 or -5, efficiently deacetylate MEF2D, thus contributing to MEF2D-dependent transcriptional repression. Our MEF2 reporter gene data are in agreement with these findings, excluding HDAC4 catalytic activity for being responsible for transcriptional repression and pointing to the involvement of class I enzymes that are equally sensitive to the inhibition by both LAQ824 and apicidin.
Besides their transcriptional corepressor functions, a number of “atypical” biological activities have been described for different class IIa members. It has been reported that HDAC7 localizes to mitochondria and may regulate the initiation of apoptosis (30). HDAC7 was also shown to bind HIF-1α and activate transcription (31). More recently, HDAC4, together with HDAC6, was also found to interact with and stabilize HIF-1α (32). A role for HDAC4 in the DNA damage-response pathway was proposed based on colocalization with 53BP1 nuclear foci after exposure to agents causing double-stranded DNA breaks (33). Furthermore, independent findings showed that the hydroxamate LBH589 confines HDAC4 to the cytoplasm and increases the duration of γ-H2AX foci in irradiated cells (34). However, the precise role of HDAC4 catalytic activity in these biological contexts remains to be elucidated.
Finally, our data also imply that class IIa HDACs should be considered a distinct class of therapeutic targets, concerning both their natural substrates and selective mechanisms of inhibition. Future work will undoubtedly identify these yet unknown physiological substrates and further reveal the already complex biological activities of these enzymes.
Methods
Sequence Analysis and Modeling.
Class IIa HDAC sequences for C. intestinalis, C. savignyi, and H. roretzi were assembled by using data from the National Center for Biotechnology Information nucleotide sequence database (see legend to SI Fig. 7). Sequences were aligned with CLUSTALW followed by manual refinement with Genedoc (www.psc.edu/biomed/genedoc). Graphical display and structural analysis of the bacterial class II homolog (Protein Data Bank entry 1ZZ1, www.pdb.org) was performed by using ViewerLite (Accelrys, San Diego).
Deacetylase Assays.
The histone and peptide activity assays were performed by using 3[H]acetyl-core histones purified from HeLa cells as described in refs. 35 and 36 or a fluorescent histone H4 (AcK16) peptide substrate [pepH4-AcK16: Mca-GAK(ε-Ac)RHRKV-NH2; AnaSpec, San Jose, CA]. Enzymes were incubated in 100 μl of assay buffer (50 mM Hepes, pH 7.4/5% Glycerol/0.01% Triton X-100/0.1 mg/ml BSA) containing 5–8 μg (7,000–10,000 dpm) of 3[H]acetyl-histones or 4 μM pepH4-AcK16. Reactions were performed at room temperature (RT) for 4 h and terminated with 20 μl of a 0.5 M HCl/1 M acetic acid mixture (histones), or 10% TFA (peptide). 3[H]acetic acid released from histones was extracted with 1 ml ethyl acetate, 900 μl of solvent phase containing free 3[H]acetic acid and 90 μl of aqueous phase containing unprocessed 3[H]acetyl-histones were collected, and radioactivity was measured by scintillation counting. The percentage conversion was calculated as: [(dpm product/dpm product + dpm substrate) × 100]. The percentage of pepH4-AcK16 converted to the corresponding deacetylated product was determined by HPLC on a Beckman C18 column (Beckman Instruments, Columbia, MD) eluted with a linear gradient of acetonitrile in H2O. The HDAC fluorescent activity assay (BIOMOL Research Laboratories, Plymouth Meeting, PA) was performed in 96-well plates according to the manufacturer's instructions by using either the commercial Fluor de Lys substrate (KI-104) or the trifluoroacetyl-lysine substrate (for synthesis, see SI Methods).
HDACi activities were evaluated by preincubating compounds and recombinant enzymes in assay buffer at RT for 15 min, before substrate addition. Mean values of at least two independent assays are reported.
HDAC4 Compound Binding Assays.
For cross-linking experiments, 10 nM affinity-purified flag-tagged HDAC4, HDAC3, or HDAC6 were incubated under activity assay conditions with the LAQ824 derivative, the apicidin derivative, or the negative-control compound lacking the HDACi moiety. Compounds were tested at 40, 60, or 90 nM concentrations, corresponding to 1:4, 1:6, or 1:9 protein:compound ratios. After 2 h of incubation at RT, UV cross-linking was performed, and polypeptides covalently cross-linked to the biotinylated compounds were separated by SDS/PAGE and analyzed by Western blot using alkaline phosphatase-conjugated ExtrAvidin (Sigma, St. Louis, MO).
For pull-down experiments with HDAC4-FLAG, soluble extracts from HEK293 cells transiently transfected with pHDAC4-FLAG were prepared. Suitable aliquots containing 2.5 μg of HDAC4-FLAG (0.2 μM in 100 μl) were incubated for 3 h at RT with streptavidin-coated magnetic beads (Dynal Biotech, Lake Success, NY) pretreated with each of the three 5 μM biotinylated compounds also used in cross-linking experiments. Unbound proteins were collected, and bound proteins were detached from the compound-coated beads by boiling in SDS dye. Both fractions were loaded on SDS/PAGE gels and visualized by Coomassie blue staining. For competition experiments, different HDACis were added at concentrations of 50 μM to the HDAC4-FLAG-overexpressing extracts for 3 h at RT before incubation with the LAQ824-matrix. Similar conditions were used for pull-down experiments by using the recombinant HDAC4-CD purified from E. coli.
Supplementary Material
Acknowledgments
We wish to dedicate this publication to the memory of Giovanni Migliaccio, a dear friend and colleague, who has been an example of scientific rigor, integrity, and enthusiasm to all of us. We would like to acknowledge Giovanni's contribution to the HDAC project and to this publication in many lively and fruitful discussions. We thank S. Altamura, O. Cecchetti, and S. Serafini for performing some activity assays; V. Richon for careful reading of the manuscript; and M. Emili for artwork. This work was partly supported by a grant from the Italian Ministero dell' Istruzione, dell' Università e della Ricerca.
Abbreviations
- HDAC
histone deacetylase
- HDACi
histone deacetylase inhibitor
- HDAC4-CD
HDAC4 catalytic domain
- RT
room temperature.
Footnotes
Conflict of interest statement: All authors are employees of Merck, Sharp, and Dohme.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0706487104/DC1.
References
- 1.Glozak MA, Sengupta N, Zhang X, Seto E. Gene. 2005;363:15–23. doi: 10.1016/j.gene.2005.09.010. [DOI] [PubMed] [Google Scholar]
- 2.Roth SY, Denu JM, Allis CD. Annu Rev Biochem. 2001;70:81–120. doi: 10.1146/annurev.biochem.70.1.81. [DOI] [PubMed] [Google Scholar]
- 3.de Ruijter AJ, van Gennip AH, Caron HN, Kemp S, van Kuilenburg AB. Biochem J. 2003;370:737–749. doi: 10.1042/BJ20021321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Verdin E, Dequiedt F, Kasler HG. Trends Genet. 2003;19:286–293. doi: 10.1016/S0168-9525(03)00073-8. [DOI] [PubMed] [Google Scholar]
- 5.Gregoretti IV, Lee YM, Goodson HV. J Mol Biol. 2004;338:17–31. doi: 10.1016/j.jmb.2004.02.006. [DOI] [PubMed] [Google Scholar]
- 6.Blander G, Guarente L. Annu Rev Biochem. 2004;73:417–435. doi: 10.1146/annurev.biochem.73.011303.073651. [DOI] [PubMed] [Google Scholar]
- 7.Finnin MS, Donigian JR, Cohen A, Richon VM, Rifkind RA, Marks PA, Breslow R, Pavletich NP. Nature. 1999;401:188–193. doi: 10.1038/43710. [DOI] [PubMed] [Google Scholar]
- 8.Nielsen TK, Hildmann C, Dickmanns A, Schwienhorst A, Ficner R. J Mol Biol. 2005;354:107–120. doi: 10.1016/j.jmb.2005.09.065. [DOI] [PubMed] [Google Scholar]
- 9.Somoza JR, Skene RJ, Katz BA, Mol C, Ho JD, Jennings AJ, Luong C, Arvai A, Buggy JJ, Chi E, et al. Structure (London) 2004;12:1325–1334. doi: 10.1016/j.str.2004.04.012. [DOI] [PubMed] [Google Scholar]
- 10.Vannini A, Volpari C, Filocamo G, Casavola EC, Brunetti M, Renzoni D, Chakravarty P, Paolini C, De Francesco R, Gallinari P, et al. Proc Natl Acad Sci USA. 2004;101:15064–15069. doi: 10.1073/pnas.0404603101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang CL, McKinsey TA, Chang S, Antos CL, Hill JA, Olson EN. Cell. 2002;110:479–488. doi: 10.1016/s0092-8674(02)00861-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vega RB, Matsuda K, Oh J, Barbosa AC, Yang X, Meadows E, McAnally J, Pomajzl C, Shelton JM, Richardson JA, et al. Cell. 2004;119:555–566. doi: 10.1016/j.cell.2004.10.024. [DOI] [PubMed] [Google Scholar]
- 13.Chang S, McKinsey TA, Zhang CL, Richardson JA, Hill JA, Olson EN. Mol Cell Biol. 2004;24:8467–8476. doi: 10.1128/MCB.24.19.8467-8476.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dequiedt F, Kasler H, Fischle W, Kiermer V, Weinstein M, Herndier BG, Verdin E. Immunity. 2003;18:687–698. doi: 10.1016/s1074-7613(03)00109-2. [DOI] [PubMed] [Google Scholar]
- 15.Yang XJ, Gregoire S. Mol Cell Biol. 2005;25:2873–2884. doi: 10.1128/MCB.25.8.2873-2884.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhou X, Richon VM, Rifkind RA, Marks PA. Proc Natl Acad Sci USA. 2000;97:1056–1061. doi: 10.1073/pnas.97.3.1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sparrow DB, Miska EA, Langley E, Reynaud-Deonauth S, Kotecha S, Towers N, Spohr G, Kouzarides T, Mohun TJ. EMBO J. 1999;18:5085–5098. doi: 10.1093/emboj/18.18.5085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chan JK, Sun L, Yang XJ, Zhu G, Wu Z. J Biol Chem. 2003;278:23515–23521. doi: 10.1074/jbc.M301922200. [DOI] [PubMed] [Google Scholar]
- 19.Fischle W, Dequiedt F, Fillion M, Hendzel MJ, Voelter W, Verdin E. J Biol Chem. 2001;276:35826–35835. doi: 10.1074/jbc.M104935200. [DOI] [PubMed] [Google Scholar]
- 20.Fischle W, Dequiedt F, Hendzel MJ, Guenther MG, Lazar MA, Voelter W, Verdin E. Mol Cell. 2002;9:45–57. doi: 10.1016/s1097-2765(01)00429-4. [DOI] [PubMed] [Google Scholar]
- 21.Wang AH, Bertos NR, Vezmar M, Pelletier N, Crosato M, Heng HH, Th'ng J, Han J, Yang XJ. Mol Cell Biol. 1999;19:7816–7827. doi: 10.1128/mcb.19.11.7816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Guenther MG, Barak O, Lazar MA. Mol Cell Biol. 2001;21:6091–6101. doi: 10.1128/MCB.21.18.6091-6101.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang J, Kalkum M, Chait BT, Roeder RG. Mol Cell. 2002;9:611–623. doi: 10.1016/s1097-2765(02)00468-9. [DOI] [PubMed] [Google Scholar]
- 24.Hassig CA, Tong JK, Fleischer TC, Owa T, Grable PG, Ayer DE, Schreiber SL. Proc Natl Acad Sci USA. 1998;95:3519–3524. doi: 10.1073/pnas.95.7.3519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cho Y, Griswold A, Campbell C, Min KT. Genomics. 2005;86:606–617. doi: 10.1016/j.ygeno.2005.07.007. [DOI] [PubMed] [Google Scholar]
- 26.Zeremski M, Stricker JR, Fischer D, Zusman SB, Cohen D. Genesis. 2003;35:31–38. doi: 10.1002/gene.10159. [DOI] [PubMed] [Google Scholar]
- 27.Jin YH, Jeon EJ, Li QL, Lee YH, Choi JK, Kim WJ, Lee KY, Bae SC. J Biol Chem. 2004;279:29409–29417. doi: 10.1074/jbc.M313120200. [DOI] [PubMed] [Google Scholar]
- 28.Zhao X, Sternsdorf T, Bolger TA, Evans RM, Yao TP. Mol Cell Biol. 2005;25:8456–8464. doi: 10.1128/MCB.25.19.8456-8464.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gregoire S, Xiao L, Nie J, Zhang X, Xu M, Li J, Wong J, Seto E, Yang XJ. Mol Cell Biol. 2007;27:1280–1295. doi: 10.1128/MCB.00882-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bakin RE, Jung MO. J Biol Chem. 2004;279:51218–51225. doi: 10.1074/jbc.M409271200. [DOI] [PubMed] [Google Scholar]
- 31.Kato H, Tamamizu-Kato S, Shibasaki F. J Biol Chem. 2004;279:41966–41974. doi: 10.1074/jbc.M406320200. [DOI] [PubMed] [Google Scholar]
- 32.Qian DZ, Kachhap SK, Collis SJ, Verheul HM, Carducci MA, Atadja P, Pili R. Cancer Res. 2006;66:8814–8821. doi: 10.1158/0008-5472.CAN-05-4598. [DOI] [PubMed] [Google Scholar]
- 33.Kao GD, McKenna WG, Guenther MG, Muschel RJ, Lazar MA, Yen TJ. J Cell Biol. 2003;160:1017–1027. doi: 10.1083/jcb.200209065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Geng L, Cuneo KC, Fu A, Tu T, Atadja PW, Hallahan DE. Cancer Res. 2006;66:11298–11304. doi: 10.1158/0008-5472.CAN-06-0049. [DOI] [PubMed] [Google Scholar]
- 35.Yoshida M, Kijima M, Akita M, Beppu T. J Biol Chem. 1990;265:17174–17179. [PubMed] [Google Scholar]
- 36.Inoue A, Fujimoto D. Biochim Biophys Acta. 1970;220:307–316. doi: 10.1016/0005-2744(70)90015-x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.