Abstract
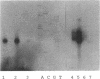
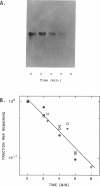
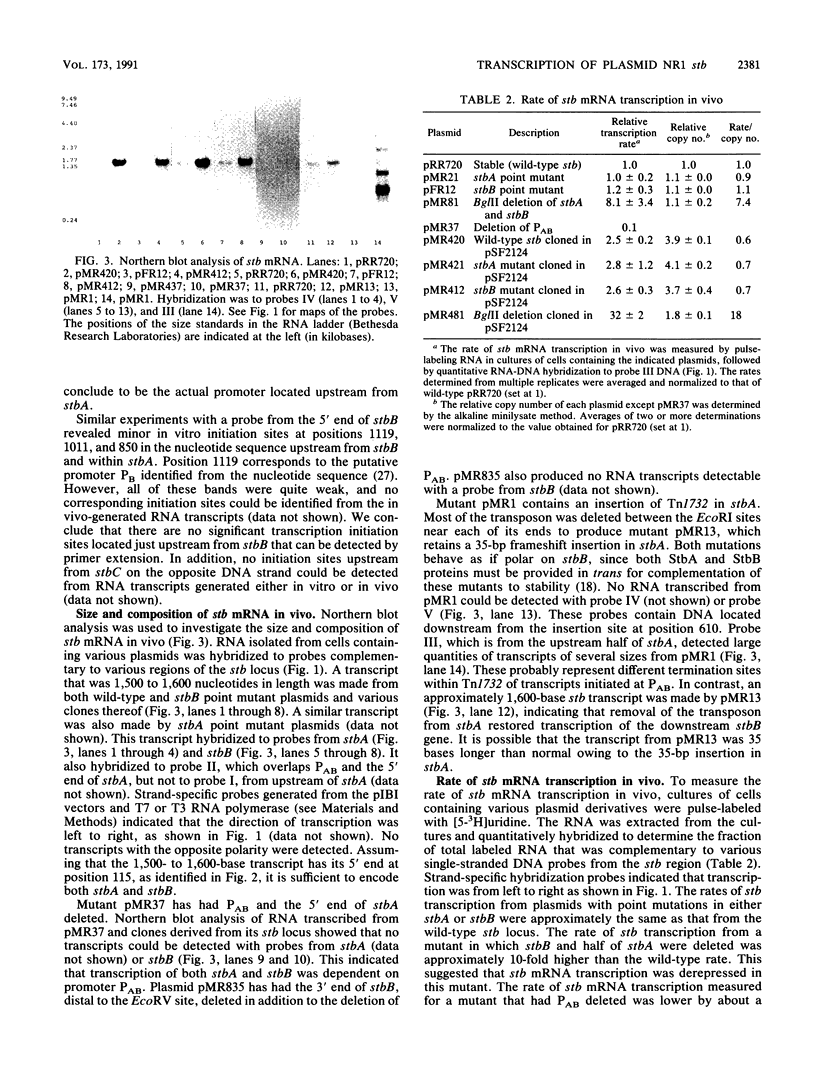
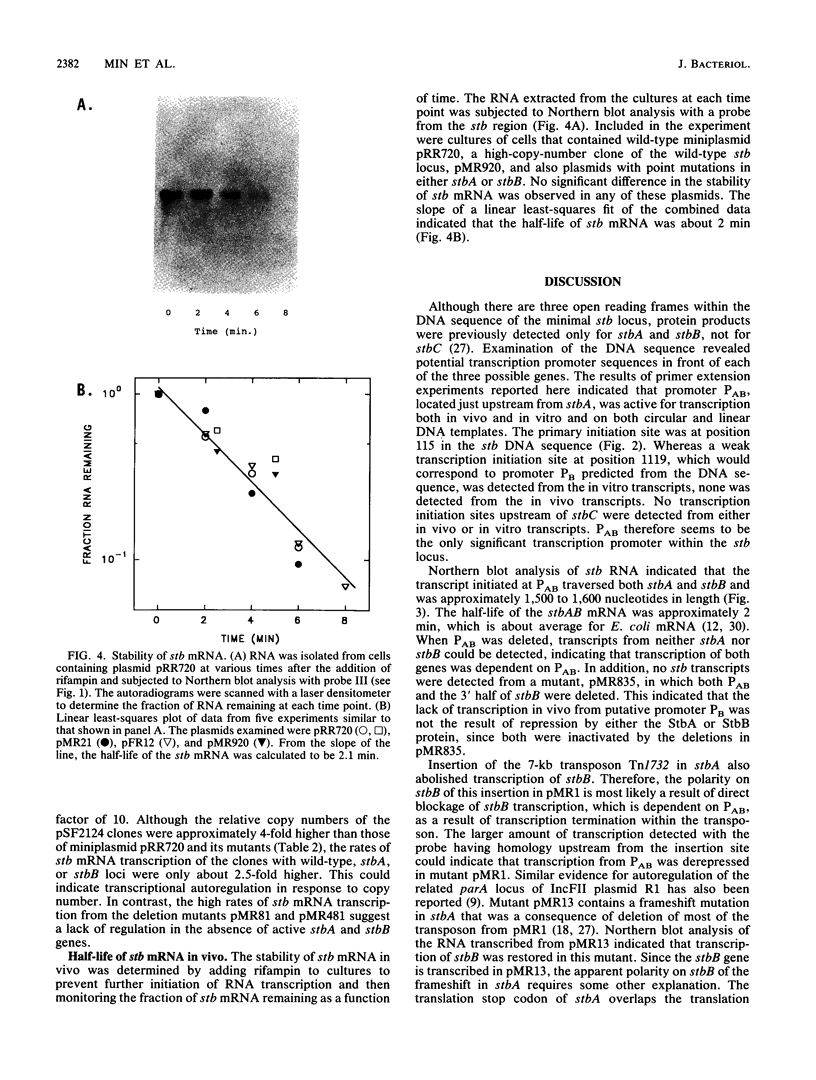
The stability (stb) locus of IncFII plasmid NR1 is composed of an essential cis-acting DNA site located upstream from two tandem genes that encode essential stability proteins. The stb locus was found to be transcribed from a promoter site just upstream from the first gene, stbA. This promoter was active for transcription both in vivo and in vitro and was located within the region that includes the essential cis-acting site. Transcripts initiated from this site were approximately 1,500 to 1,600 nucleotides in length. Northern (RNA) blot analysis indicated that the transcripts traversed both stbA and the downstream gene, stbB. Mutants from which the promoter had been deleted failed to produce detectable transcripts from either stbA or stbB. Transcription of a third open reading frame, stbC, which is contained within the stbB gene in the opposite DNA strand, could not be detected. For a mutant in which a transposon had been inserted in stbA, no transcription of stbB was detected. After deletion of most of the transposon, which left behind a 35-bp frameshift insertion in stbA, transcription of stbB was restored, although the insertion still had a polar effect on stbB function. The rate of in vivo transcription of the stb locus was measured by pulse-labeling of RNA followed by quantitative RNA-DNA hybridization. Mutants deleted of stbB had an approximately 10-fold increase in the rate of transcription, whereas those deleted of the promoter region had at least a 10-fold reduction in transcription rate. The half-life of stb mRNA was approximately 2 min. These data suggest that stbA and stbB are cotranscribed as an operon that may be autoregulated.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Abeles A. L., Friedman S. A., Austin S. J. Partition of unit-copy miniplasmids to daughter cells. III. The DNA sequence and functional organization of the P1 partition region. J Mol Biol. 1985 Sep 20;185(2):261–272. doi: 10.1016/0022-2836(85)90402-4. [DOI] [PubMed] [Google Scholar]
- Austin S. J. Plasmid partition. Plasmid. 1988 Jul;20(1):1–9. doi: 10.1016/0147-619x(88)90001-7. [DOI] [PubMed] [Google Scholar]
- Austin S., Abeles A. Partition of unit-copy miniplasmids to daughter cells. I. P1 and F miniplasmids contain discrete, interchangeable sequences sufficient to promote equipartition. J Mol Biol. 1983 Sep 15;169(2):353–372. doi: 10.1016/s0022-2836(83)80055-2. [DOI] [PubMed] [Google Scholar]
- Austin S., Abeles A. Partition of unit-copy miniplasmids to daughter cells. II. The partition region of miniplasmid P1 encodes an essential protein and a centromere-like site at which it acts. J Mol Biol. 1983 Sep 15;169(2):373–387. doi: 10.1016/s0022-2836(83)80056-4. [DOI] [PubMed] [Google Scholar]
- Austin S., Nordström K. Partition-mediated incompatibility of bacterial plasmids. Cell. 1990 Feb 9;60(3):351–354. doi: 10.1016/0092-8674(90)90584-2. [DOI] [PubMed] [Google Scholar]
- Birnboim H. C., Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979 Nov 24;7(6):1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deuschle U., Kammerer W., Gentz R., Bujard H. Promoters of Escherichia coli: a hierarchy of in vivo strength indicates alternate structures. EMBO J. 1986 Nov;5(11):2987–2994. doi: 10.1002/j.1460-2075.1986.tb04596.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerdes K., Molin S. Partitioning of plasmid R1. Structural and functional analysis of the parA locus. J Mol Biol. 1986 Aug 5;190(3):269–279. doi: 10.1016/0022-2836(86)90001-x. [DOI] [PubMed] [Google Scholar]
- Glisin V., Crkvenjakov R., Byus C. Ribonucleic acid isolated by cesium chloride centrifugation. Biochemistry. 1974 Jun 4;13(12):2633–2637. doi: 10.1021/bi00709a025. [DOI] [PubMed] [Google Scholar]
- Gold L. Posttranscriptional regulatory mechanisms in Escherichia coli. Annu Rev Biochem. 1988;57:199–233. doi: 10.1146/annurev.bi.57.070188.001215. [DOI] [PubMed] [Google Scholar]
- Kennell D., Riezman H. Transcription and translation initiation frequencies of the Escherichia coli lac operon. J Mol Biol. 1977 Jul;114(1):1–21. doi: 10.1016/0022-2836(77)90279-0. [DOI] [PubMed] [Google Scholar]
- Kornblum J. S., Projan S. J., Moghazeh S. L., Novick R. P. A rapid method to quantitate non-labeled RNA species in bacterial cells. Gene. 1988;63(1):75–85. doi: 10.1016/0378-1119(88)90547-1. [DOI] [PubMed] [Google Scholar]
- Lau L. F., Roberts J. W. Rho-dependent transcription termination at lambda R1 requires upstream sequences. J Biol Chem. 1985 Jan 10;260(1):574–584. [PubMed] [Google Scholar]
- Miki T., Easton A. M., Rownd R. H. Cloning of replication, incompatibility, and stability functions of R plasmid NR1. J Bacteriol. 1980 Jan;141(1):87–99. doi: 10.1128/jb.141.1.87-99.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miki T., Easton A. M., Rownd R. H. Mapping of the resistance genes of the R plasmid NR1. Mol Gen Genet. 1978 Jan 17;158(3):217–224. doi: 10.1007/BF00267192. [DOI] [PubMed] [Google Scholar]
- Min Y. N., Tabuchi A., Fan Y. L., Womble D. D., Rownd R. H. Complementation of mutants of the stability locus of IncFII plasmid NR1. Essential functions of the trans-acting stbA and stbB gene products. J Mol Biol. 1988 Nov 20;204(2):345–356. doi: 10.1016/0022-2836(88)90581-5. [DOI] [PubMed] [Google Scholar]
- NAKAYA R., NAKAMURA A., MURATA Y. Resistance transfer agents in Shigella. Biochem Biophys Res Commun. 1960 Dec;3:654–659. doi: 10.1016/0006-291x(60)90081-4. [DOI] [PubMed] [Google Scholar]
- Nordström K., Austin S. J. Mechanisms that contribute to the stable segregation of plasmids. Annu Rev Genet. 1989;23:37–69. doi: 10.1146/annurev.ge.23.120189.000345. [DOI] [PubMed] [Google Scholar]
- Ogura T., Hiraga S. Partition mechanism of F plasmid: two plasmid gene-encoded products and a cis-acting region are involved in partition. Cell. 1983 Feb;32(2):351–360. doi: 10.1016/0092-8674(83)90454-3. [DOI] [PubMed] [Google Scholar]
- Peterson B. C., Rownd R. H. Recombination sites in plasmid drug resistance gene amplification. J Bacteriol. 1985 Dec;164(3):1359–1361. doi: 10.1128/jb.164.3.1359-1361.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rownd R., Nakaya R., Nakamura A. Molecular nature of the drug-resistance factors of the Enterobacteriaceae. J Mol Biol. 1966 Jun;17(2):376–393. doi: 10.1016/s0022-2836(66)80149-3. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- So M., Gill R., Falkow S. The generation of a ColE1-Apr cloning vehicle which allows detection of inserted DNA. Mol Gen Genet. 1975 Dec 30;142(3):239–249. doi: 10.1007/BF00425649. [DOI] [PubMed] [Google Scholar]
- Tabuchi A., Min Y. N., Kim C. K., Fan Y. L., Womble D. D., Rownd R. H. Genetic organization and nucleotide sequence of the stability locus of IncFII plasmid NR1. J Mol Biol. 1988 Aug 5;202(3):511–525. doi: 10.1016/0022-2836(88)90282-3. [DOI] [PubMed] [Google Scholar]
- Ubben D., Schmitt R. Tn1721 derivatives for transposon mutagenesis, restriction mapping and nucleotide sequence analysis. Gene. 1986;41(2-3):145–152. doi: 10.1016/0378-1119(86)90093-4. [DOI] [PubMed] [Google Scholar]
- Vieira J., Messing J. The pUC plasmids, an M13mp7-derived system for insertion mutagenesis and sequencing with synthetic universal primers. Gene. 1982 Oct;19(3):259–268. doi: 10.1016/0378-1119(82)90015-4. [DOI] [PubMed] [Google Scholar]
- Womble D. D., Rownd R. H. Genetic and physical map of plasmid NR1: comparison with other IncFII antibiotic resistance plasmids. Microbiol Rev. 1988 Dec;52(4):433–451. doi: 10.1128/mr.52.4.433-451.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Womble D. D., Sampathkumar P., Easton A. M., Luckow V. A., Rownd R. H. Transcription of the replication control region of the IncFII R-plasmid NR1 in vitro and in vivo. J Mol Biol. 1985 Feb 5;181(3):395–410. doi: 10.1016/0022-2836(85)90228-1. [DOI] [PubMed] [Google Scholar]
- Yanisch-Perron C., Vieira J., Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33(1):103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- von Gabain A., Belasco J. G., Schottel J. L., Chang A. C., Cohen S. N. Decay of mRNA in Escherichia coli: investigation of the fate of specific segments of transcripts. Proc Natl Acad Sci U S A. 1983 Feb;80(3):653–657. doi: 10.1073/pnas.80.3.653. [DOI] [PMC free article] [PubMed] [Google Scholar]