Abstract
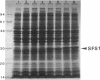
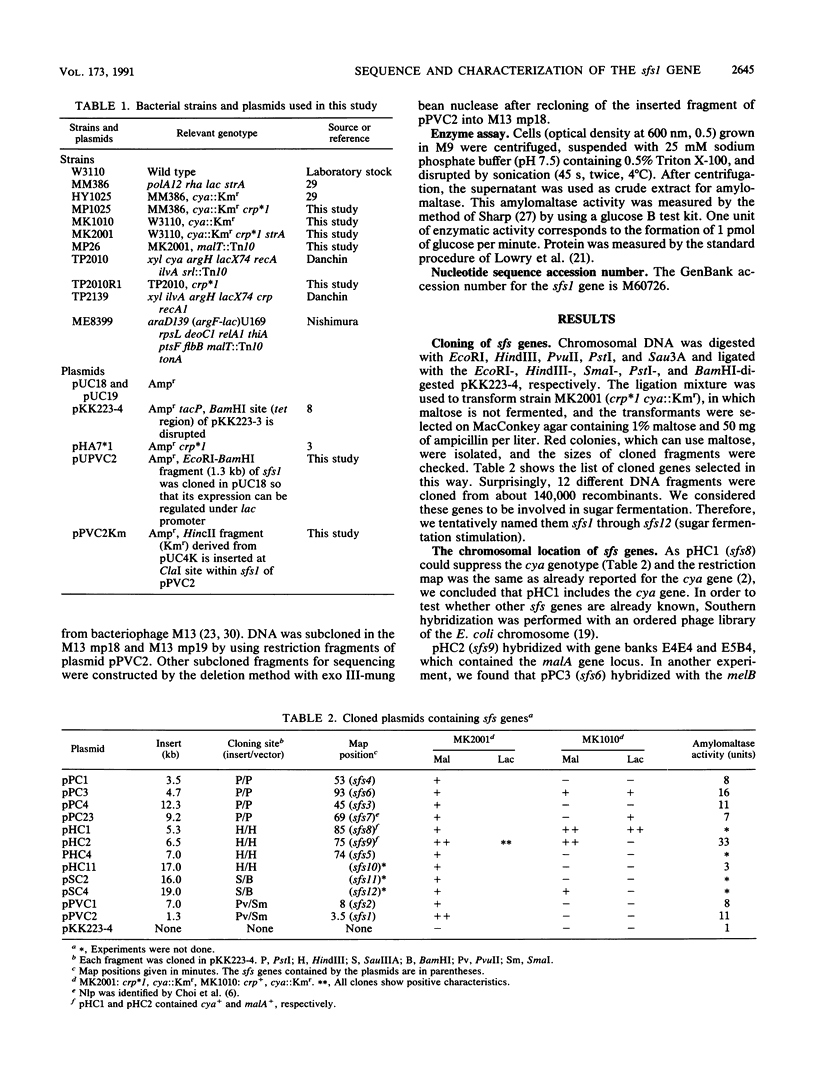
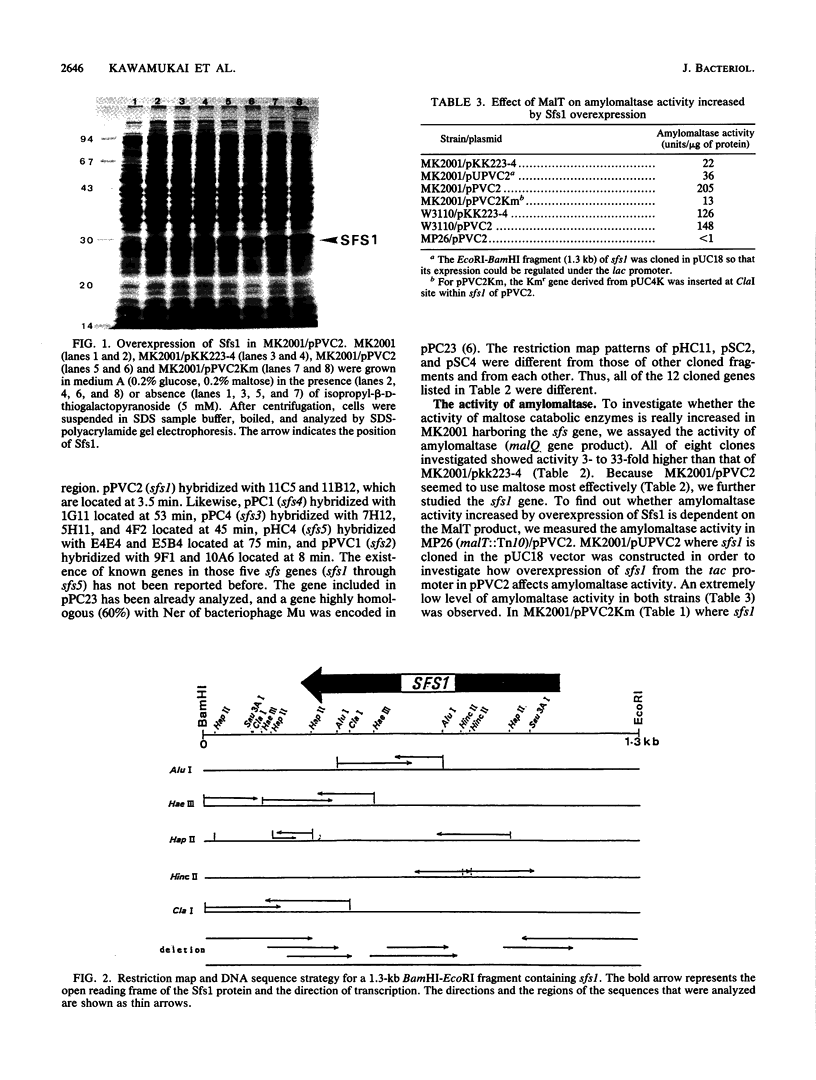
We have cloned at least 12 different Escherichia coli genes which enable strain MK2001 to use maltose. The genes were designated sfs1 through sfs12 (sugar fermentation stimulation). Previously, one (sfs7) of them was mapped at 65 min on the E. coli chromosome and identified as nlp, which has high homology to repressor protein (Ner) of Mu phage, which contains a putative DNA binding region (Y.-L. Choi, T. Nishida, M. Kawamukai, R. Utsumi, H. Sakai, and T. Komano, J. Bacteriol. 171:5222-5225, 1989). In this study, another gene (sfs1) located at 3.5 min was newly found and analyzed. The nucleotide sequence of sfs1 encoded a protein of 234 amino acids (molecular mass, 26,227 Da) which also has a putative DNA binding domain. Overexpression of the sfs1 gene in MK2001 resulted in a 10-fold increase of amylomaltase, which was still dependent on MalT. These results suggest that Sfs1 could be a new regulatory factor involved in maltose metabolism.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Adhya S., Garges S. How cyclic AMP and its receptor protein act in Escherichia coli. Cell. 1982 Jun;29(2):287–289. doi: 10.1016/0092-8674(82)90145-3. [DOI] [PubMed] [Google Scholar]
- Aiba H., Kawamukai M., Ishihama A. Cloning and promoter analysis of the Escherichia coli adenylate cyclase gene. Nucleic Acids Res. 1983 Jun 11;11(11):3451–3465. doi: 10.1093/nar/11.11.3451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aiba H., Nakamura T., Mitani H., Mori H. Mutations that alter the allosteric nature of cAMP receptor protein of Escherichia coli. EMBO J. 1985 Dec 1;4(12):3329–3332. doi: 10.1002/j.1460-2075.1985.tb04084.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birnboim H. C., Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979 Nov 24;7(6):1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennan R. G., Matthews B. W. The helix-turn-helix DNA binding motif. J Biol Chem. 1989 Feb 5;264(4):1903–1906. [PubMed] [Google Scholar]
- Choi Y. L., Nishida T., Kawamukai M., Utsumi R., Sakai H., Komano T. Cloning and sequencing of an Escherichia coli gene, nlp, highly homologous to the ner genes of bacteriophages Mu and D108. J Bacteriol. 1989 Sep;171(9):5222–5225. doi: 10.1128/jb.171.9.5222-5225.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garges S., Adhya S. Sites of allosteric shift in the structure of the cyclic AMP receptor protein. Cell. 1985 Jul;41(3):745–751. doi: 10.1016/s0092-8674(85)80055-6. [DOI] [PubMed] [Google Scholar]
- Gent M. E., Gronenborn A. M., Davies R. W., Clore G. M. Probing the sequence-specific interaction of the cyclic AMP receptor protein with DNA by site-directed mutagenesis. Biochem J. 1987 Mar 15;242(3):645–653. doi: 10.1042/bj2420645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goad W. B., Kanehisa M. I. Pattern recognition in nucleic acid sequences. I. A general method for finding local homologies and symmetries. Nucleic Acids Res. 1982 Jan 11;10(1):247–263. doi: 10.1093/nar/10.1.247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gronenborn A. M., Sandulache R., Gärtner S., Clore G. M. Mutations in the cyclic AMP binding site of the cyclic AMP receptor protein of Escherichia coli. Biochem J. 1988 Aug 1;253(3):801–807. doi: 10.1042/bj2530801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harman J. G., McKenney K., Peterkofsky A. Structure-function analysis of three cAMP-independent forms of the cAMP receptor protein. J Biol Chem. 1986 Dec 15;261(35):16332–16339. [PubMed] [Google Scholar]
- Harman J. G., Peterkofsky A., McKenney K. Arginine substituted for leucine at position 195 produces a cyclic AMP-independent form of the Escherichia coli cyclic AMP receptor protein. J Biol Chem. 1988 Jun 15;263(17):8072–8077. [PubMed] [Google Scholar]
- Hu J. C., Gross C. A. Mutations in rpoD that increase expression of genes in the mal regulon of Escherichia coli K-12. J Mol Biol. 1988 Sep 5;203(1):15–27. doi: 10.1016/0022-2836(88)90087-3. [DOI] [PubMed] [Google Scholar]
- Kang P. J., Craig E. A. Identification and characterization of a new Escherichia coli gene that is a dosage-dependent suppressor of a dnaK deletion mutation. J Bacteriol. 1990 Apr;172(4):2055–2064. doi: 10.1128/jb.172.4.2055-2064.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawamukai M., Kishimoto J., Utsumi R., Himeno M., Komano T., Aiba H. Negative regulation of adenylate cyclase gene (cya) expression by cyclic AMP-cyclic AMP receptor protein in Escherichia coli: studies with cya-lac protein and operon fusion plasmids. J Bacteriol. 1985 Nov;164(2):872–877. doi: 10.1128/jb.164.2.872-877.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohara Y., Akiyama K., Isono K. The physical map of the whole E. coli chromosome: application of a new strategy for rapid analysis and sorting of a large genomic library. Cell. 1987 Jul 31;50(3):495–508. doi: 10.1016/0092-8674(87)90503-4. [DOI] [PubMed] [Google Scholar]
- Kyte J., Doolittle R. F. A simple method for displaying the hydropathic character of a protein. J Mol Biol. 1982 May 5;157(1):105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- LOWRY O. H., ROSEBROUGH N. J., FARR A. L., RANDALL R. J. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951 Nov;193(1):265–275. [PubMed] [Google Scholar]
- Messing J. New M13 vectors for cloning. Methods Enzymol. 1983;101:20–78. doi: 10.1016/0076-6879(83)01005-8. [DOI] [PubMed] [Google Scholar]
- Raibaud O., Vidal-Ingigliardi D., Richet E. A complex nucleoprotein structure involved in activation of transcription of two divergent Escherichia coli promoters. J Mol Biol. 1989 Feb 5;205(3):471–485. doi: 10.1016/0022-2836(89)90218-0. [DOI] [PubMed] [Google Scholar]
- Sharp P. Interference in glucose oxidase-peroxidase blood glucose methods. Clin Chim Acta. 1972 Aug;40(1):115–120. doi: 10.1016/0009-8981(72)90257-4. [DOI] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Yanisch-Perron C., Vieira J., Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33(1):103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- de Boer H. A., Comstock L. J., Vasser M. The tac promoter: a functional hybrid derived from the trp and lac promoters. Proc Natl Acad Sci U S A. 1983 Jan;80(1):21–25. doi: 10.1073/pnas.80.1.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Crombrugghe B., Busby S., Buc H. Cyclic AMP receptor protein: role in transcription activation. Science. 1984 May 25;224(4651):831–838. doi: 10.1126/science.6372090. [DOI] [PubMed] [Google Scholar]