Abstract
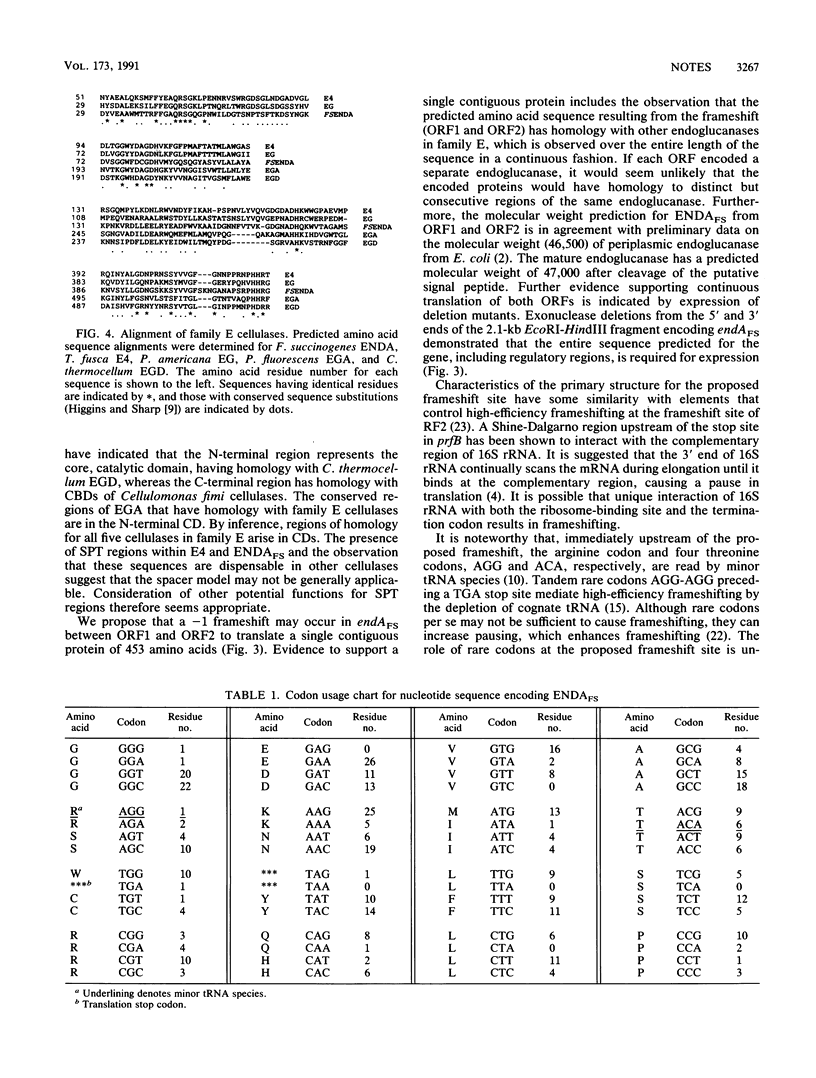
The complete nucleotide sequence of endAFS, an endoglucanase gene isolated from the ruminal anaerobe Fibrobacter succinogenes AR1, was determined. endAFS encodes two overlapping open reading frames (ORF1 and ORF2), and it was proposed that a -1 ribosomal frameshift was required to allow contiguous synthesis of a 453-amino-acid endoglucanase. A proline- and threonine-rich region at the C terminus of ORF1 and rare codons for arginine and threonine were coincident with the proposed frameshift site. ENDAFS is proposed to be a member of subgroup 1 of family E endoglucanases, of which endoglucanases from Thermomonospora fusca and Persea americana (avocado) are also members. Endoglucanases from Clostridium thermocellum and Pseudomonas fluorescens form subgroup 2.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Cavicchioli R., Watson K. Molecular cloning, expression, and characterization of endoglucanase genes from Fibrobacter succinogenes AR1. Appl Environ Microbiol. 1991 Feb;57(2):359–365. doi: 10.1128/aem.57.2.359-365.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craigen W. J., Lee C. C., Caskey C. T. Recent advances in peptide chain termination. Mol Microbiol. 1990 Jun;4(6):861–865. doi: 10.1111/j.1365-2958.1990.tb00658.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira L. M., Durrant A. J., Hall J., Hazlewood G. P., Gilbert H. J. Spatial separation of protein domains is not necessary for catalytic activity or substrate binding in a xylanase. Biochem J. 1990 Jul 1;269(1):261–264. doi: 10.1042/bj2690261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall J., Gilbert H. J. The nucleotide sequence of a carboxymethylcellulase gene from Pseudomonas fluorescens subsp. cellulosa. Mol Gen Genet. 1988 Jul;213(1):112–117. doi: 10.1007/BF00333406. [DOI] [PubMed] [Google Scholar]
- Hartz D., McPheeters D. S., Traut R., Gold L. Extension inhibition analysis of translation initiation complexes. Methods Enzymol. 1988;164:419–425. doi: 10.1016/s0076-6879(88)64058-4. [DOI] [PubMed] [Google Scholar]
- Henrissat B., Claeyssens M., Tomme P., Lemesle L., Mornon J. P. Cellulase families revealed by hydrophobic cluster analysis. Gene. 1989 Sep 1;81(1):83–95. doi: 10.1016/0378-1119(89)90339-9. [DOI] [PubMed] [Google Scholar]
- Higgins D. G., Sharp P. M. CLUSTAL: a package for performing multiple sequence alignment on a microcomputer. Gene. 1988 Dec 15;73(1):237–244. doi: 10.1016/0378-1119(88)90330-7. [DOI] [PubMed] [Google Scholar]
- Ikemura T. Correlation between the abundance of Escherichia coli transfer RNAs and the occurrence of the respective codons in its protein genes: a proposal for a synonymous codon choice that is optimal for the E. coli translational system. J Mol Biol. 1981 Sep 25;151(3):389–409. doi: 10.1016/0022-2836(81)90003-6. [DOI] [PubMed] [Google Scholar]
- Joliff G., Béguin P., Aubert J. P. Nucleotide sequence of the cellulase gene celD encoding endoglucanase D of Clostridium thermocellum. Nucleic Acids Res. 1986 Nov 11;14(21):8605–8613. doi: 10.1093/nar/14.21.8605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGavin M. J., Forsberg C. W., Crosby B., Bell A. W., Dignard D., Thomas D. Y. Structure of the cel-3 gene from Fibrobacter succinogenes S85 and characteristics of the encoded gene product, endoglucanase 3. J Bacteriol. 1989 Oct;171(10):5587–5595. doi: 10.1128/jb.171.10.5587-5595.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spanjaard R. A., van Duin J. Translation of the sequence AGG-AGG yields 50% ribosomal frameshift. Proc Natl Acad Sci U S A. 1988 Nov;85(21):7967–7971. doi: 10.1073/pnas.85.21.7967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teather R. M., Erfle J. D. DNA sequence of a Fibrobacter succinogenes mixed-linkage beta-glucanase (1,3-1,4-beta-D-glucan 4-glucanohydrolase) gene. J Bacteriol. 1990 Jul;172(7):3837–3841. doi: 10.1128/jb.172.7.3837-3841.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss R. B., Dunn D. M., Dahlberg A. E., Atkins J. F., Gesteland R. F. Reading frame switch caused by base-pair formation between the 3' end of 16S rRNA and the mRNA during elongation of protein synthesis in Escherichia coli. EMBO J. 1988 May;7(5):1503–1507. doi: 10.1002/j.1460-2075.1988.tb02969.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss R. B., Gallant J. A. Frameshift suppression in aminoacyl-tRNA limited cells. Genetics. 1986 Apr;112(4):727–739. doi: 10.1093/genetics/112.4.727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Heijne G. Transcending the impenetrable: how proteins come to terms with membranes. Biochim Biophys Acta. 1988 Jun 9;947(2):307–333. doi: 10.1016/0304-4157(88)90013-5. [DOI] [PubMed] [Google Scholar]