Abstract
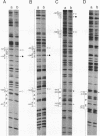
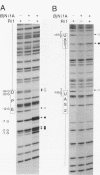
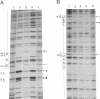
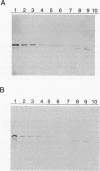
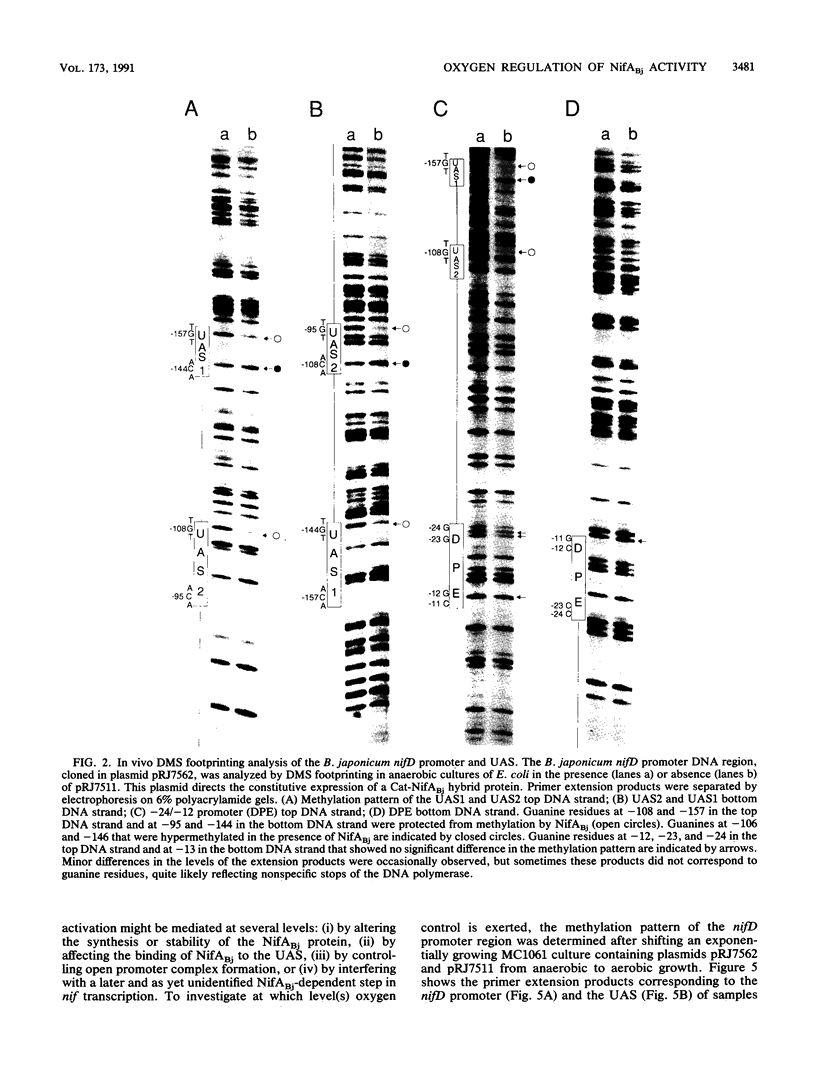
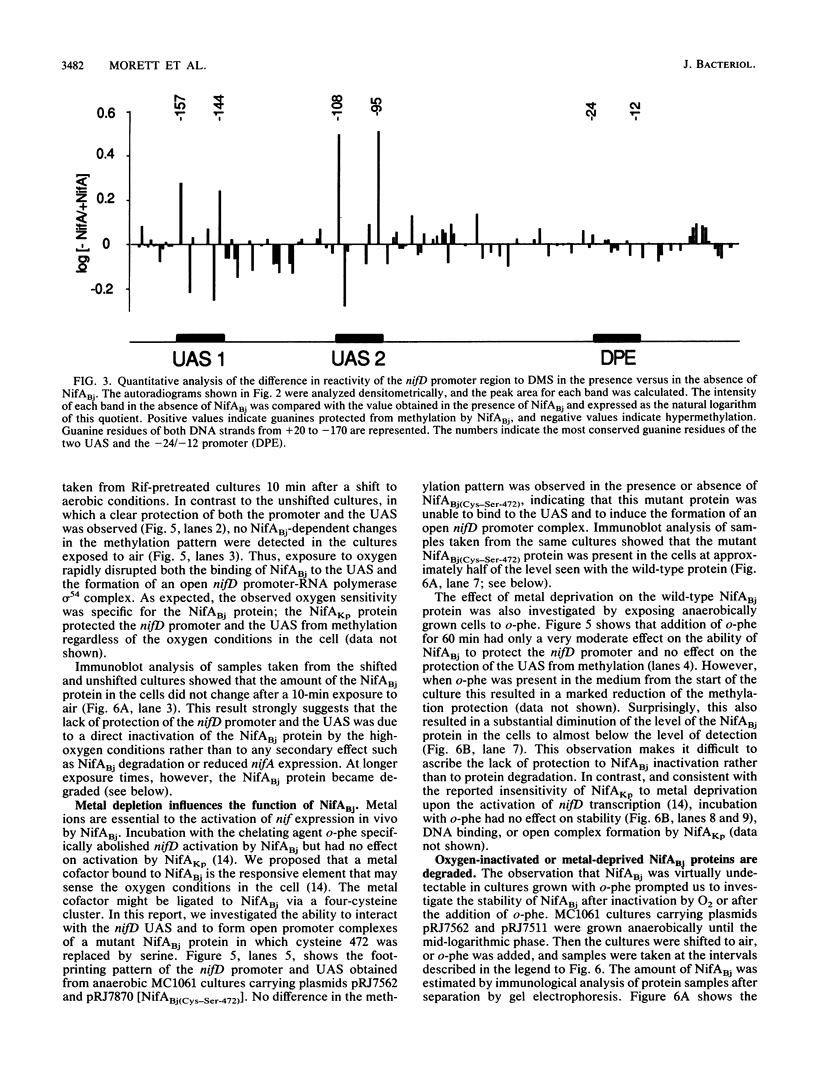
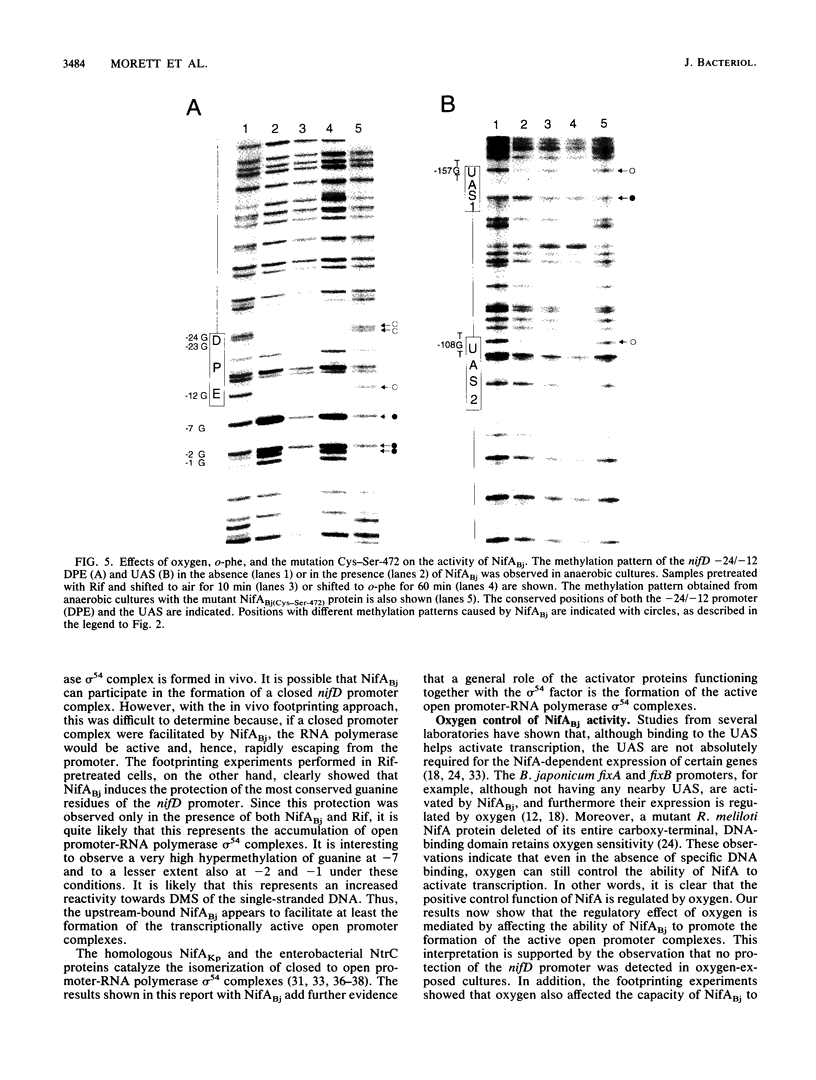
Central to the genetic regulatory circuit that controls Bradyrhizobium japonicum nif and fix gene expression is the NifA protein. NifA activates transcription of several nif and fix genes and autoregulates its expression during symbiosis in soybean root nodules or in free-living microaerobic conditions. High O2 tensions result in the lack of nif expression, possibly by inactivation of NifA through oxidation of an essential metal cofactor. Several B. japonicum nif and fix promoters have upstream activator sequences (UAS) required for optimal activation. The UAS are located more than 100 bp from the -24/-12 promoter and have been proposed to be binding sites for NifA. We investigated the interaction of NifA with the nifD promoter region by using in vivo dimethyl sulfate footprinting. NifA-dependent protection from methylation of the two UAS of this promoter was detected. Footprinting experiments in the presence of rifampin showed that UAS-bound NifA led to the formation of an open nifD promoter-RNA polymerase sigma 54 complex. Shift to aerobic growth resulted in a rapid loss of protection of both the UAS and the promoter, indicating that the DNA-binding and the activation functions of NifA were controlled by the O2 status of the cell. After an almost complete inactivation by oxygen, the NifA protein began to degrade. Furthermore, metal deprivation also caused degradation of NifA. In this case, however, the rates of NifA inactivation and NifA degradation were not clearly distinguishable. The results are discussed in the light of a previously proposed model, according to which the oxidation state of a NifA-metal complex influences the conformation of NifA for both DNA-binding and positive control functions.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Alvarez-Morales A., Betancourt-Alvarez M., Kaluza K., Hennecke H. Activation of the Bradyrhizobium japonicum nifH and nifDK operons is dependent on promoter-upstream DNA sequences. Nucleic Acids Res. 1986 May 27;14(10):4207–4227. doi: 10.1093/nar/14.10.4207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anthamatten D., Hennecke H. The regulatory status of the fixL- and fixJ-like genes in Bradyrhizobium japonicum may be different from that in Rhizobium meliloti. Mol Gen Genet. 1991 Jan;225(1):38–48. doi: 10.1007/BF00282640. [DOI] [PubMed] [Google Scholar]
- Beynon J. L., Williams M. K., Cannon F. C. Expression and functional analysis of the Rhizobium meliloti nifA gene. EMBO J. 1988 Jan;7(1):7–14. doi: 10.1002/j.1460-2075.1988.tb02777.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borowiec J. A., Gralla J. D. High-resolution analysis of lac transcription complexes inside cells. Biochemistry. 1986 Sep 9;25(18):5051–5057. doi: 10.1021/bi00366a012. [DOI] [PubMed] [Google Scholar]
- Buchanan-Wollaston V., Cannon M. C., Beynon J. L., Cannon F. C. Role of the nifA gene product in the regulation of nif expression in Klebsiella pneumoniae. Nature. 1981 Dec 24;294(5843):776–778. doi: 10.1038/294776a0. [DOI] [PubMed] [Google Scholar]
- Buck M., Cannon W., Woodcock J. Transcriptional activation of the Klebsiella pneumoniae nitrogenase promoter may involve DNA loop formation. Mol Microbiol. 1987 Sep;1(2):243–249. doi: 10.1111/j.1365-2958.1987.tb00518.x. [DOI] [PubMed] [Google Scholar]
- Cannon W. V., Kreutzer R., Kent H. M., Morett E., Buck M. Activation of the Klebsiella pneumoniae nifU promoter: identification of multiple and overlapping upstream NifA binding sites. Nucleic Acids Res. 1990 Apr 11;18(7):1693–1701. doi: 10.1093/nar/18.7.1693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casadaban M. J., Cohen S. N. Analysis of gene control signals by DNA fusion and cloning in Escherichia coli. J Mol Biol. 1980 Apr;138(2):179–207. doi: 10.1016/0022-2836(80)90283-1. [DOI] [PubMed] [Google Scholar]
- Dixon R., Kennedy C., Kondorosi A., Krishnapillai V., Merrick M. Complementation analysis of Klebsiella pneumoniae mutants defective in nitrogen fixation. Mol Gen Genet. 1977 Nov 29;157(2):189–198. doi: 10.1007/BF00267397. [DOI] [PubMed] [Google Scholar]
- Drummond M., Whitty P., Wootton J. Sequence and domain relationships of ntrC and nifA from Klebsiella pneumoniae: homologies to other regulatory proteins. EMBO J. 1986 Feb;5(2):441–447. doi: 10.1002/j.1460-2075.1986.tb04230.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer H M, Hennecke H. Direct response of Bradyrhizobium japonicum nifA-mediated nif gene regulation to cellular oxygen status. Mol Gen Genet. 1987 Oct;209(3):621–626. doi: 10.1007/BF00331174. [DOI] [PubMed] [Google Scholar]
- Fischer H. M., Bruderer T., Hennecke H. Essential and non-essential domains in the Bradyrhizobium japonicum NifA protein: identification of indispensable cysteine residues potentially involved in redox reactivity and/or metal binding. Nucleic Acids Res. 1988 Mar 25;16(5):2207–2224. doi: 10.1093/nar/16.5.2207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer H. M., Fritsche S., Herzog B., Hennecke H. Critical spacing between two essential cysteine residues in the interdomain linker of the Bradyrhizobium japonicum NifA protein. FEBS Lett. 1989 Sep 11;255(1):167–171. doi: 10.1016/0014-5793(89)81083-x. [DOI] [PubMed] [Google Scholar]
- Gottesman S. Genetics of proteolysis in Escherichia coli*. Annu Rev Genet. 1989;23:163–198. doi: 10.1146/annurev.ge.23.120189.001115. [DOI] [PubMed] [Google Scholar]
- Gubler M. Fine-tuning of nif and fix gene expression by upstream activator sequences in Bradyrhizobium japonicum. Mol Microbiol. 1989 Feb;3(2):149–159. doi: 10.1111/j.1365-2958.1989.tb01804.x. [DOI] [PubMed] [Google Scholar]
- Gubler M., Hennecke H. Regulation of the fixA gene and fixBC operon in Bradyrhizobium japonicum. J Bacteriol. 1988 Mar;170(3):1205–1214. doi: 10.1128/jb.170.3.1205-1214.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gussin G. N., Ronson C. W., Ausubel F. M. Regulation of nitrogen fixation genes. Annu Rev Genet. 1986;20:567–591. doi: 10.1146/annurev.ge.20.120186.003031. [DOI] [PubMed] [Google Scholar]
- Hennecke H. Nitrogen fixation genes involved in the Bradyrhizobium japonicum-soybean symbiosis. FEBS Lett. 1990 Aug 1;268(2):422–426. doi: 10.1016/0014-5793(90)81297-2. [DOI] [PubMed] [Google Scholar]
- Hertig C., Li R. Y., Louarn A. M., Garnerone A. M., David M., Batut J., Kahn D., Boistard P. Rhizobium meliloti regulatory gene fixJ activates transcription of R. meliloti nifA and fixK genes in Escherichia coli. J Bacteriol. 1989 Mar;171(3):1736–1738. doi: 10.1128/jb.171.3.1736-1738.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoover T. R., Santero E., Porter S., Kustu S. The integration host factor stimulates interaction of RNA polymerase with NIFA, the transcriptional activator for nitrogen fixation operons. Cell. 1990 Oct 5;63(1):11–22. doi: 10.1016/0092-8674(90)90284-l. [DOI] [PubMed] [Google Scholar]
- Huala E., Ausubel F. M. The central domain of Rhizobium meliloti NifA is sufficient to activate transcription from the R. meliloti nifH promoter. J Bacteriol. 1989 Jun;171(6):3354–3365. doi: 10.1128/jb.171.6.3354-3365.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huala E., Moon A. L., Ausubel F. M. Aerobic inactivation of Rhizobium meliloti NifA in Escherichia coli is mediated by lon and two newly identified genes, snoB and snoC. J Bacteriol. 1991 Jan;173(1):382–390. doi: 10.1128/jb.173.1.382-390.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kullik I., Fritsche S., Knobel H., Sanjuan J., Hennecke H., Fischer H. M. Bradyrhizobium japonicum has two differentially regulated, functional homologs of the sigma 54 gene (rpoN). J Bacteriol. 1991 Feb;173(3):1125–1138. doi: 10.1128/jb.173.3.1125-1138.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kustu S., Santero E., Keener J., Popham D., Weiss D. Expression of sigma 54 (ntrA)-dependent genes is probably united by a common mechanism. Microbiol Rev. 1989 Sep;53(3):367–376. doi: 10.1128/mr.53.3.367-376.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minchin S. D., Austin S., Dixon R. A. Transcriptional activation of the Klebsiella pneumoniae nifLA promoter by NTRC is face-of-the-helix dependent and the activator stabilizes the interaction of sigma 54-RNA polymerase with the promoter. EMBO J. 1989 Nov;8(11):3491–3499. doi: 10.1002/j.1460-2075.1989.tb08514.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morett E., Buck M. In vivo studies on the interaction of RNA polymerase-sigma 54 with the Klebsiella pneumoniae and Rhizobium meliloti nifH promoters. The role of NifA in the formation of an open promoter complex. J Mol Biol. 1989 Nov 5;210(1):65–77. doi: 10.1016/0022-2836(89)90291-x. [DOI] [PubMed] [Google Scholar]
- Morett E., Buck M. NifA-dependent in vivo protection demonstrates that the upstream activator sequence of nif promoters is a protein binding site. Proc Natl Acad Sci U S A. 1988 Dec;85(24):9401–9405. doi: 10.1073/pnas.85.24.9401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morett E., Cannon W., Buck M. The DNA-binding domain of the transcriptional activator protein NifA resides in its carboxy terminus, recognises the upstream activator sequences of nif promoters and can be separated from the positive control function of NifA. Nucleic Acids Res. 1988 Dec 23;16(24):11469–11488. doi: 10.1093/nar/16.24.11469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morett E., Kreutzer R., Cannon W., Buck M. The influence of the Klebsiella pneumoniae regulatory gene nifL upon the transcriptional activator protein NifA. Mol Microbiol. 1990 Aug;4(8):1253–1258. doi: 10.1111/j.1365-2958.1990.tb00704.x. [DOI] [PubMed] [Google Scholar]
- Ninfa A. J., Reitzer L. J., Magasanik B. Initiation of transcription at the bacterial glnAp2 promoter by purified E. coli components is facilitated by enhancers. Cell. 1987 Sep 25;50(7):1039–1046. doi: 10.1016/0092-8674(87)90170-x. [DOI] [PubMed] [Google Scholar]
- Popham D. L., Szeto D., Keener J., Kustu S. Function of a bacterial activator protein that binds to transcriptional enhancers. Science. 1989 Feb 3;243(4891):629–635. doi: 10.1126/science.2563595. [DOI] [PubMed] [Google Scholar]
- Sasse-Dwight S., Gralla J. D. Probing the Escherichia coli glnALG upstream activation mechanism in vivo. Proc Natl Acad Sci U S A. 1988 Dec;85(23):8934–8938. doi: 10.1073/pnas.85.23.8934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spiro S., Guest J. R. FNR and its role in oxygen-regulated gene expression in Escherichia coli. FEMS Microbiol Rev. 1990 Aug;6(4):399–428. doi: 10.1111/j.1574-6968.1990.tb04109.x. [DOI] [PubMed] [Google Scholar]
- Spiro S., Roberts R. E., Guest J. R. FNR-dependent repression of the ndh gene of Escherichia coli and metal ion requirement for FNR-regulated gene expression. Mol Microbiol. 1989 May;3(5):601–608. doi: 10.1111/j.1365-2958.1989.tb00207.x. [DOI] [PubMed] [Google Scholar]
- Thöny B., Anthamatten D., Hennecke H. Dual control of the Bradyrhizobium japonicum symbiotic nitrogen fixation regulatory operon fixR nifA: analysis of cis- and trans-acting elements. J Bacteriol. 1989 Aug;171(8):4162–4169. doi: 10.1128/jb.171.8.4162-4169.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thöny B., Fischer H. M., Anthamatten D., Bruderer T., Hennecke H. The symbiotic nitrogen fixation regulatory operon (fixRnifA) of Bradyrhizobium japonicum is expressed aerobically and is subject to a novel, nifA-independent type of activation. Nucleic Acids Res. 1987 Oct 26;15(20):8479–8499. doi: 10.1093/nar/15.20.8479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thöny B., Hennecke H. The -24/-12 promoter comes of age. FEMS Microbiol Rev. 1989 Dec;5(4):341–357. doi: 10.1016/0168-6445(89)90028-4. [DOI] [PubMed] [Google Scholar]
- Trageser M., Unden G. Role of cysteine residues and of metal ions in the regulatory functioning of FNR, the transcriptional regulator of anaerobic respiration in Escherichia coli. Mol Microbiol. 1989 May;3(5):593–599. doi: 10.1111/j.1365-2958.1989.tb00206.x. [DOI] [PubMed] [Google Scholar]
- Virts E. L., Stanfield S. W., Helinski D. R., Ditta G. S. Common regulatory elements control symbiotic and microaerobic induction of nifA in Rhizobium meliloti. Proc Natl Acad Sci U S A. 1988 May;85(9):3062–3065. doi: 10.1073/pnas.85.9.3062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Philip P., Batut J., Boistard P. Rhizobium meliloti Fix L is an oxygen sensor and regulates R. meliloti nifA and fixK genes differently in Escherichia coli. J Bacteriol. 1990 Aug;172(8):4255–4262. doi: 10.1128/jb.172.8.4255-4262.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]