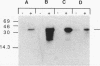
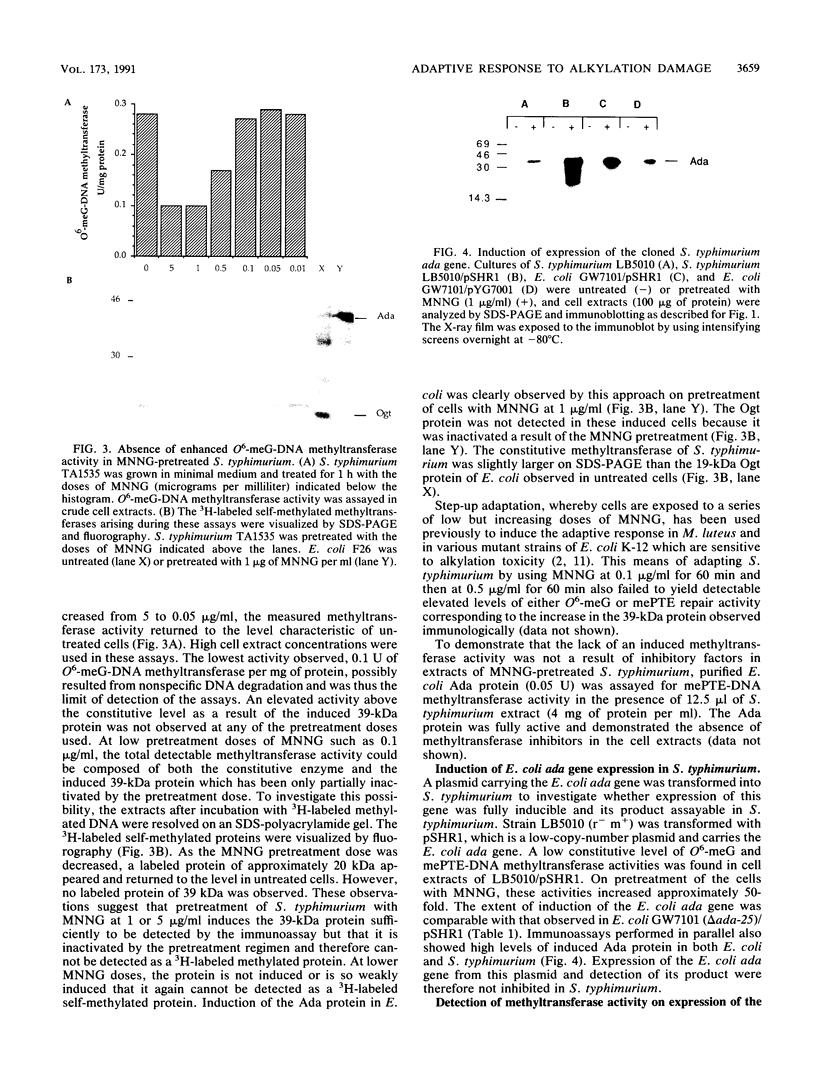
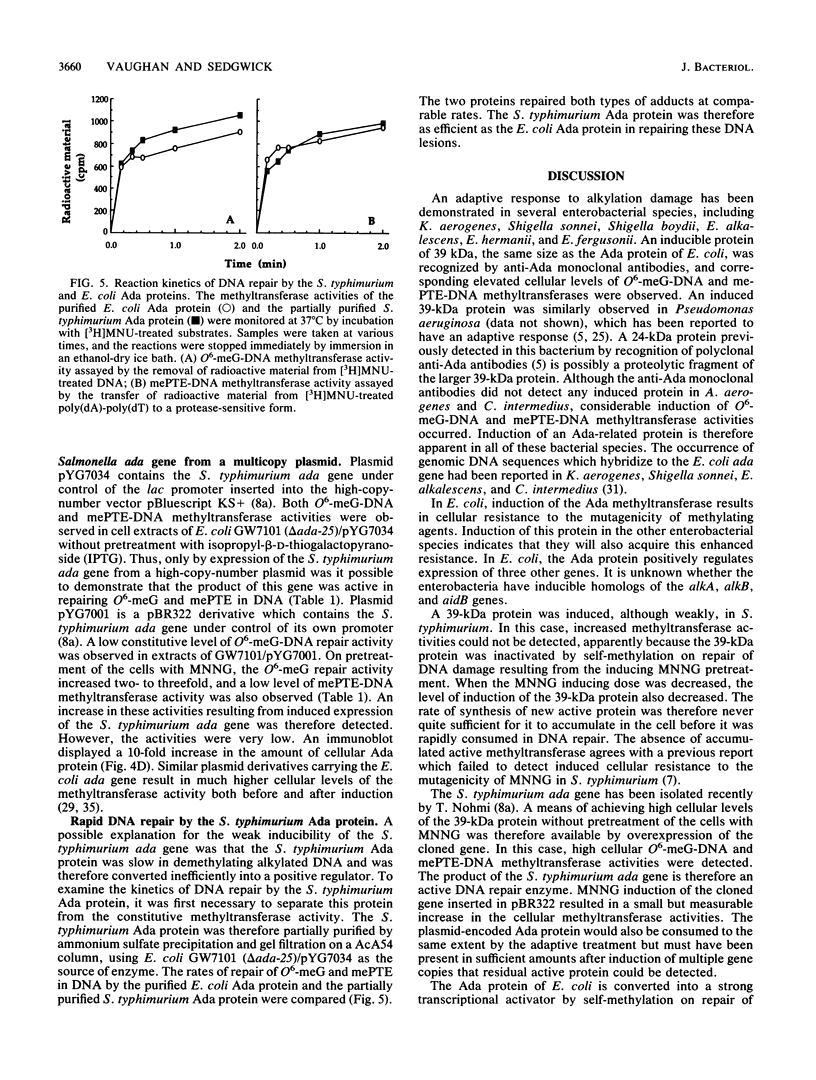
Abstract
An efficient adaptive response to alkylation damage was observed in several enterobacterial species, including Klebsiella aerogenes, Shigella sonnei, Shigella boydii, Escherichia alkalescens, Escherichia hermanii, and Escherichia fergusonii. Increased O6-methylguanine-DNA and methylphosphotriester-DNA methyltransferase activities correlated with the induction of a 39-kDa protein recognized by monoclonal antibodies raised against the Escherichia coli Ada protein. Induced methyltransferase activities were similarly observed in Aerobacter aerogenes and Citrobacter intermedius, although no antigenically cross-reacting material was present. Weak induction of a 39-kDa protein immunologically related to the E. coli Ada protein occurred in Salmonella typhimurium. This protein encoded by the cloned S. typhimurium ada gene was shown to be an active methyltransferase which repaired O6-methylguanine and methylphosphotriesters in DNA as efficiently as did the E. coli Ada protein. However, the mehtyltransferase activity of the weakly induced 39-kDa protein in S. typhimurium was not detected, apparently because it was self-methylated and thus inactivated during the adaptive N-methyl-N-nitro-N-nitrosoguanidine pretreatment. In contrast, the E. coli ada gene on a low-copy-number plasmid was efficiently induced in S. typhimurium, and high methyltransferase activities were observed. We concluded that the inefficient induction of the adaptive response in S. typhimurium results from weak transcriptional activation of its ada gene by the self-methylated protein.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ames B. N., Mccann J., Yamasaki E. Methods for detecting carcinogens and mutagens with the Salmonella/mammalian-microsome mutagenicity test. Mutat Res. 1975 Dec;31(6):347–364. doi: 10.1016/0165-1161(75)90046-1. [DOI] [PubMed] [Google Scholar]
- Ather A., Ahmed Z., Riazuddin S. Adaptive response of Micrococcus luteus to alkylating chemicals. Nucleic Acids Res. 1984 Feb 24;12(4):2111–2126. doi: 10.1093/nar/12.4.2111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrows L. R., Magee P. N. Nonenzymatic methylation of DNA by S-adenosylmethionine in vitro. Carcinogenesis. 1982;3(3):349–351. doi: 10.1093/carcin/3.3.349. [DOI] [PubMed] [Google Scholar]
- Bullas L. R., Ryu J. I. Salmonella typhimurium LT2 strains which are r- m+ for all three chromosomally located systems of DNA restriction and modification. J Bacteriol. 1983 Oct;156(1):471–474. doi: 10.1128/jb.156.1.471-474.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ceccoli J., Rosales N., Goldstein M., Yarosh D. B. Polyclonal antibodies against O6-methylguanine-DNA methyltransferase in adapted bacteria. Mutat Res. 1988 Nov;194(3):219–226. doi: 10.1016/0167-8817(88)90023-5. [DOI] [PubMed] [Google Scholar]
- Chen J., Derfler B., Samson L. Saccharomyces cerevisiae 3-methyladenine DNA glycosylase has homology to the AlkA glycosylase of E. coli and is induced in response to DNA alkylation damage. EMBO J. 1990 Dec;9(13):4569–4575. doi: 10.1002/j.1460-2075.1990.tb07910.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttenplan J. B. Comutagenic effects exerted by N-nitroso compounds. Mutat Res. 1979 Jan;66(1):25–32. doi: 10.1016/0165-1218(79)90004-1. [DOI] [PubMed] [Google Scholar]
- Guttenplan J. B., Milstein S. Resistance of Salmonella typhimurium TA 1535 to O6-guanine methylation and mutagenesis induced by low doses of N-methyl-N'-nitro-N-nitrosoguanidine: an apparent constitutive repair activity. Carcinogenesis. 1982;3(3):327–331. doi: 10.1093/carcin/3.3.327. [DOI] [PubMed] [Google Scholar]
- Hakura A., Morimoto K., Sofuni T., Nohmi T. Cloning and characterization of the Salmonella typhimurium ada gene, which encodes O6-methylguanine-DNA methyltransferase. J Bacteriol. 1991 Jun;173(12):3663–3672. doi: 10.1128/jb.173.12.3663-3672.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooley P., Shawcross S. G., Strike P. An adaptive response to alkylating agents in Aspergillus nidulans. Curr Genet. 1988 Nov;14(5):445–449. doi: 10.1007/BF00521267. [DOI] [PubMed] [Google Scholar]
- Hughes S. J., Sedgwick B. The adaptive response to alkylation damage. Constitutive expression through a mutation in the coding region of the ada gene. J Biol Chem. 1989 Dec 15;264(35):21369–21375. [PubMed] [Google Scholar]
- Jeggo P. Isolation and characterization of Escherichia coli K-12 mutants unable to induce the adaptive response to simple alkylating agents. J Bacteriol. 1979 Sep;139(3):783–791. doi: 10.1128/jb.139.3.783-791.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karran P., Lindahl T., Griffin B. Adaptive response to alkylating agents involves alteration in situ of O6-methylguanine residues in DNA. Nature. 1979 Jul 5;280(5717):76–77. doi: 10.1038/280076a0. [DOI] [PubMed] [Google Scholar]
- Kimball R. F. Further studies on the induction of mutation in Haemophilus influenzae by N-methyl-N'-nitro-N-nitrosoguanidine: lack of an inducible error-free repair system and the effect of exposure medium. Mutat Res. 1980 Aug;72(3):361–372. doi: 10.1016/0027-5107(80)90111-6. [DOI] [PubMed] [Google Scholar]
- Lindahl T., Demple B., Robins P. Suicide inactivation of the E. coli O6-methylguanine-DNA methyltransferase. EMBO J. 1982;1(11):1359–1363. doi: 10.1002/j.1460-2075.1982.tb01323.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindahl T., Sedgwick B., Sekiguchi M., Nakabeppu Y. Regulation and expression of the adaptive response to alkylating agents. Annu Rev Biochem. 1988;57:133–157. doi: 10.1146/annurev.bi.57.070188.001025. [DOI] [PubMed] [Google Scholar]
- Maga J. A., McEntee K. Response of S. cerevisiae to N-methyl-N'-nitro-N-nitrosoguanidine: mutagenesis, survival and DDR gene expression. Mol Gen Genet. 1985;200(2):313–321. doi: 10.1007/BF00425442. [DOI] [PubMed] [Google Scholar]
- Margison G. P., Cooper D. P., Potter P. M. The E. coli ogt gene. Mutat Res. 1990 Nov-Dec;233(1-2):15–21. doi: 10.1016/0027-5107(90)90146-u. [DOI] [PubMed] [Google Scholar]
- McCarthy T. V., Lindahl T. Methyl phosphotriesters in alkylated DNA are repaired by the Ada regulatory protein of E. coli. Nucleic Acids Res. 1985 Apr 25;13(8):2683–2698. doi: 10.1093/nar/13.8.2683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morohoshi F., Hayashi K., Munakata N. Bacillus subtilis ada operon encodes two DNA alkyltransferases. Nucleic Acids Res. 1990 Sep 25;18(18):5473–5480. doi: 10.1093/nar/18.18.5473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polakowska R., Perozzi G., Prakash L. Alkylation mutagenesis in Saccharomyces cerevisiae: lack of evidence for an adaptive response. Curr Genet. 1986;10(9):647–655. doi: 10.1007/BF00410912. [DOI] [PubMed] [Google Scholar]
- Potter P. M., Kleibl K., Cawkwell L., Margison G. P. Expression of the ogt gene in wild-type and ada mutants of E. coli. Nucleic Acids Res. 1989 Oct 25;17(20):8047–8060. doi: 10.1093/nar/17.20.8047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potter P. M., Wilkinson M. C., Fitton J., Carr F. J., Brennand J., Cooper D. P., Margison G. P. Characterisation and nucleotide sequence of ogt, the O6-alkylguanine-DNA-alkyltransferase gene of E. coli. Nucleic Acids Res. 1987 Nov 25;15(22):9177–9193. doi: 10.1093/nar/15.22.9177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasool S. A., Mirza A., Khan M. A. Nitrosoguanidine-induced adaptive repair in Pseudomonas aeruginosa. Curr Genet. 1990 May;17(5):417–419. doi: 10.1007/BF00334521. [DOI] [PubMed] [Google Scholar]
- Rebeck G. W., Smith C. M., Goad D. L., Samson L. Characterization of the major DNA repair methyltransferase activity in unadapted Escherichia coli and identification of a similar activity in Salmonella typhimurium. J Bacteriol. 1989 Sep;171(9):4563–4568. doi: 10.1128/jb.171.9.4563-4568.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riazuddin S., Athar A., Sohail A. Methyl transferases induced during chemical adaptation of M. luteus. Nucleic Acids Res. 1987 Nov 25;15(22):9471–9486. doi: 10.1093/nar/15.22.9471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rydberg B., Lindahl T. Nonenzymatic methylation of DNA by the intracellular methyl group donor S-adenosyl-L-methionine is a potentially mutagenic reaction. EMBO J. 1982;1(2):211–216. doi: 10.1002/j.1460-2075.1982.tb01149.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sedgwick B. Molecular cloning of a gene which regulates the adaptive response to alkylating agents in Escherichia coli. Mol Gen Genet. 1983;191(3):466–472. doi: 10.1007/BF00425764. [DOI] [PubMed] [Google Scholar]
- Sedgwick B., Robins P., Totty N., Lindahl T. Functional domains and methyl acceptor sites of the Escherichia coli ada protein. J Biol Chem. 1988 Mar 25;263(9):4430–4433. [PubMed] [Google Scholar]
- Sedgwick S. G., Goodwin P. A. Differences in mutagenic and recombinational DNA repair in enterobacteria. Proc Natl Acad Sci U S A. 1985 Jun;82(12):4172–4176. doi: 10.1073/pnas.82.12.4172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shevell D. E., Abou-Zamzam A. M., Demple B., Walker G. C. Construction of an Escherichia coli K-12 ada deletion by gene replacement in a recD strain reveals a second methyltransferase that repairs alkylated DNA. J Bacteriol. 1988 Jul;170(7):3294–3296. doi: 10.1128/jb.170.7.3294-3296.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shevell D. E., Friedman B. M., Walker G. C. Resistance to alkylation damage in Escherichia coli: role of the Ada protein in induction of the adaptive response. Mutat Res. 1990 Nov-Dec;233(1-2):53–72. doi: 10.1016/0027-5107(90)90151-s. [DOI] [PubMed] [Google Scholar]
- Takano K., Nakabeppu Y., Sekiguchi M. Functional sites of the Ada regulatory protein of Escherichia coli. Analysis by amino acid substitutions. J Mol Biol. 1988 May 20;201(2):261–271. doi: 10.1016/0022-2836(88)90137-4. [DOI] [PubMed] [Google Scholar]
- Teo I., Sedgwick B., Demple B., Li B., Lindahl T. Induction of resistance to alkylating agents in E. coli: the ada+ gene product serves both as a regulatory protein and as an enzyme for repair of mutagenic damage. EMBO J. 1984 Sep;3(9):2151–2157. doi: 10.1002/j.1460-2075.1984.tb02105.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teo I., Sedgwick B., Kilpatrick M. W., McCarthy T. V., Lindahl T. The intracellular signal for induction of resistance to alkylating agents in E. coli. Cell. 1986 Apr 25;45(2):315–324. doi: 10.1016/0092-8674(86)90396-x. [DOI] [PubMed] [Google Scholar]
- Vaughan P., Sedgwick B., Hall J., Gannon J., Lindahl T. Environmental mutagens that induce the adaptive response to alkylating agents in Escherichia coli. Carcinogenesis. 1991 Feb;12(2):263–268. doi: 10.1093/carcin/12.2.263. [DOI] [PubMed] [Google Scholar]