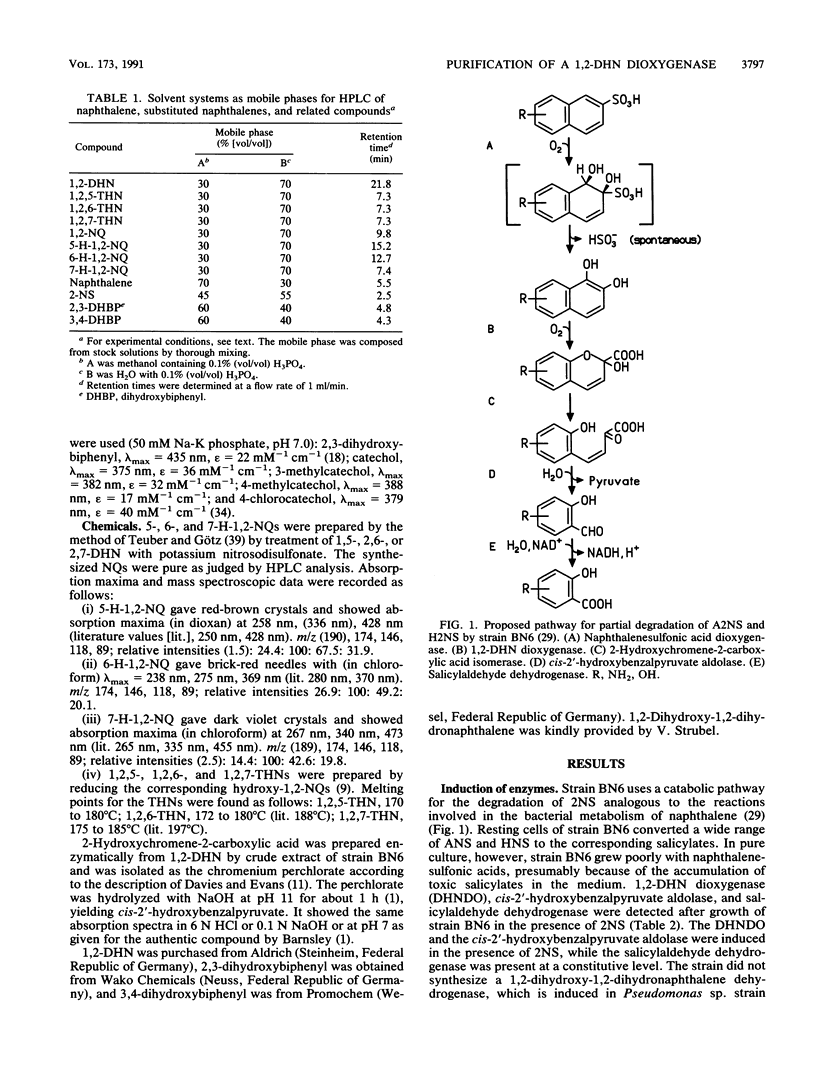
Abstract
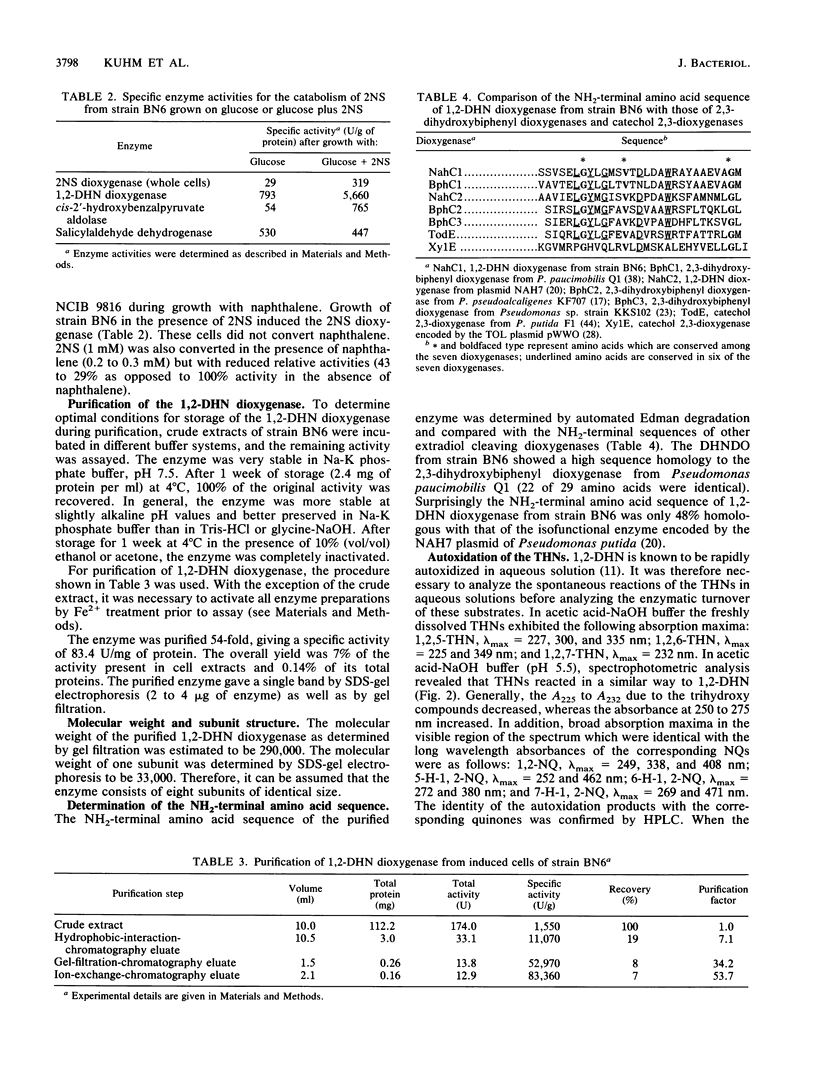
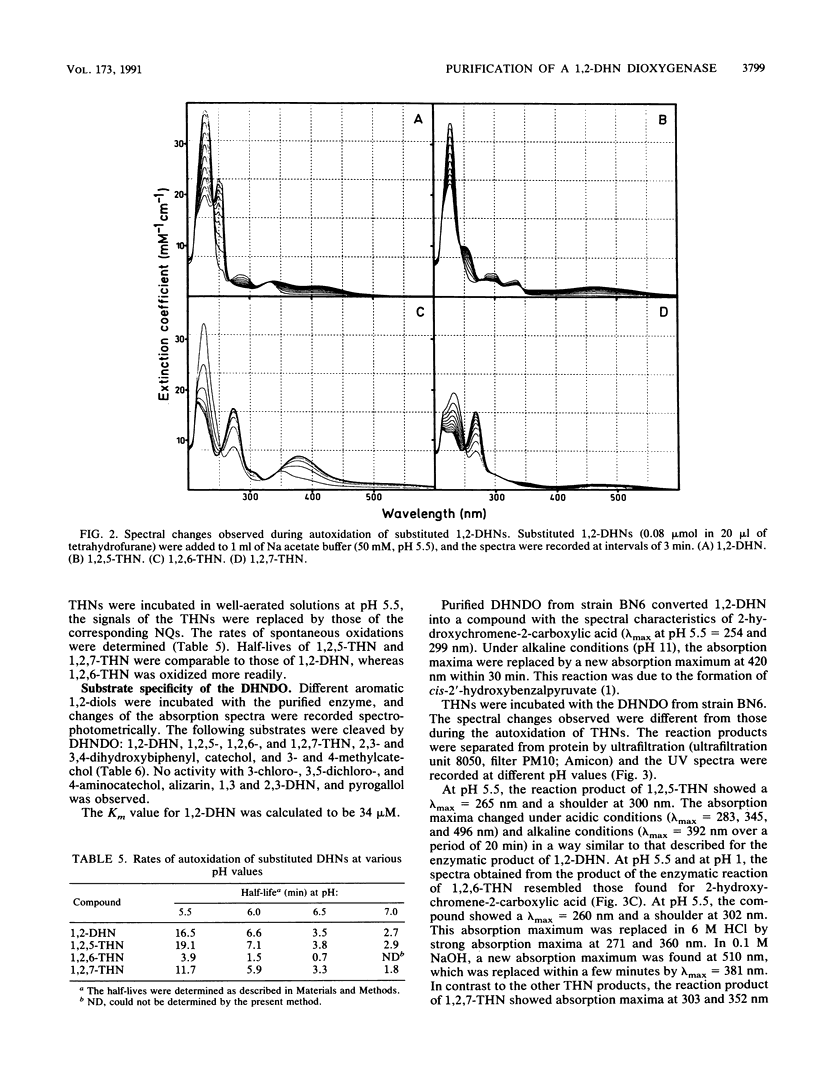
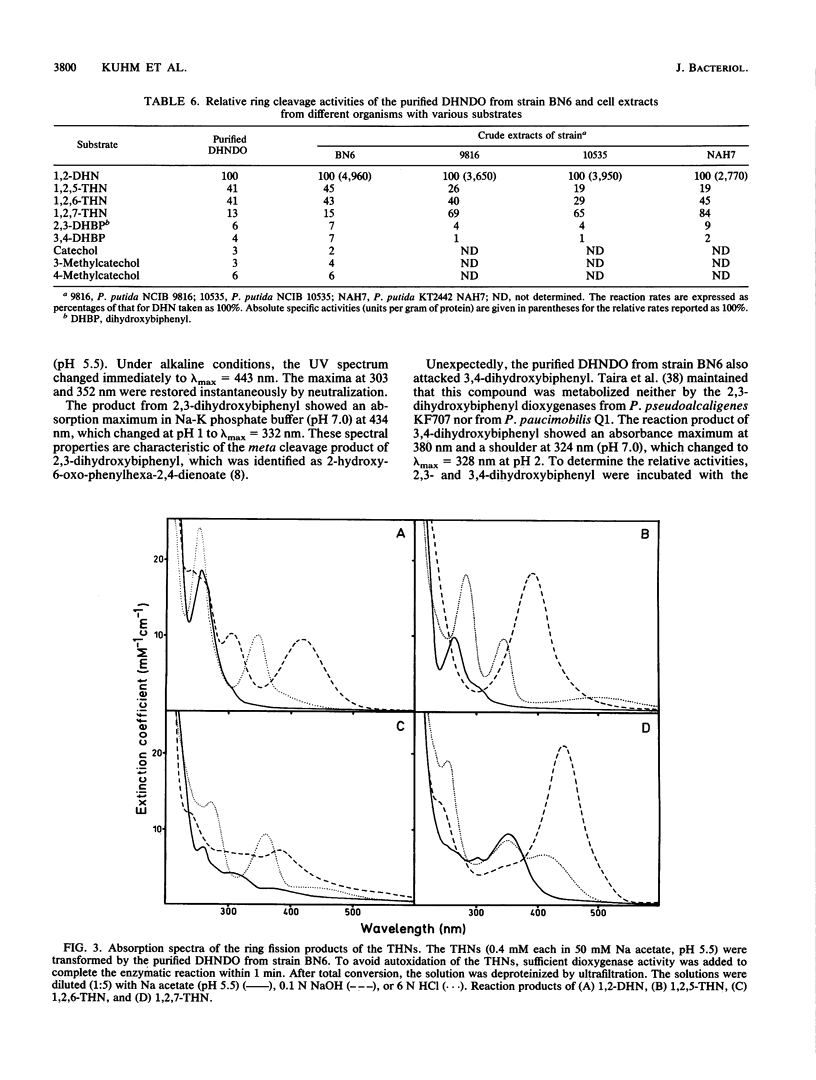
1,2-Dihydroxynaphthalene dioxygenase was purified to homogeneity from a bacterium that degrades naphthalenesulfonic acids (strain BN6). The enzyme requires Fe2+ for maximal activity and consists of eight identical subunits with a molecular weight of about 33,000. Analysis of the NH2-terminal amino acid sequence revealed a high degree of homology (22 of 29 amino acids) with the NH2-terminal amino acid sequence of 2,3-dihydroxybiphenyl dioxygenase from strain Pseudomonas paucimobilis Q1. 1,2-Dihydroxynaphthalene dioxygenase from strain BN6 shows a wide substrate specificity and also cleaves 5-, 6-, and 7-hydroxy-1,2-dihydroxynaphthalene, 2,3- and 3,4-dihydroxybiphenyl, catechol, and 3-methyl- and 4-methylcatechol. Similar activities against the hydroxy-1,2-dihydroxynaphthalenes were also found in cell extracts from naphthalene-degrading bacteria.
Full text
PDF



Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Barnsley E. A. Naphthalene metabolism by pseudomonads: the oxidation of 1,2-dihydroxynaphthalene to 2-hydroxychromene-2-carboxylic acid and the formation of 2'-hydroxybenzalpyruvate. Biochem Biophys Res Commun. 1976 Oct 4;72(3):1116–1121. doi: 10.1016/s0006-291x(76)80247-1. [DOI] [PubMed] [Google Scholar]
- Bradford M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976 May 7;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Brilon C., Beckmann W., Hellwig M., Knackmuss H. J. Enrichment and isolation of naphthalenesulfonic Acid-utilizing pseudomonads. Appl Environ Microbiol. 1981 Jul;42(1):39–43. doi: 10.1128/aem.42.1.39-43.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brilon C., Beckmann W., Knackmuss H. J. Catabolism of Naphthalenesulfonic Acids by Pseudomonas sp. A3 and Pseudomonas sp. C22. Appl Environ Microbiol. 1981 Jul;42(1):44–55. doi: 10.1128/aem.42.1.44-55.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catelani D., Colombi A. Metabolism of biphenyl. Structure and physicochemical properties of 2-hydroxy-6-oxo-6-phenylhexa-2,4-dienoic acid, the meta-cleavage product from 2,3-dihydroxybiphenyl by Pseudomonas putida. Biochem J. 1974 Nov;143(2):431–434. doi: 10.1042/bj1430431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connors M. A., Barnsley E. A. Naphthalene plasmids in pseudomonads. J Bacteriol. 1982 Mar;149(3):1096–1101. doi: 10.1128/jb.149.3.1096-1101.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies J. I., Evans W. C. Oxidative metabolism of naphthalene by soil pseudomonads. The ring-fission mechanism. Biochem J. 1964 May;91(2):251–261. doi: 10.1042/bj0910251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorn E., Hellwig M., Reineke W., Knackmuss H. J. Isolation and characterization of a 3-chlorobenzoate degrading pseudomonad. Arch Microbiol. 1974;99(1):61–70. doi: 10.1007/BF00696222. [DOI] [PubMed] [Google Scholar]
- Dunn N. W., Gunsalus I. C. Transmissible plasmid coding early enzymes of naphthalene oxidation in Pseudomonas putida. J Bacteriol. 1973 Jun;114(3):974–979. doi: 10.1128/jb.114.3.974-979.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckhardt T. A rapid method for the identification of plasmid desoxyribonucleic acid in bacteria. Plasmid. 1978 Sep;1(4):584–588. doi: 10.1016/0147-619x(78)90016-1. [DOI] [PubMed] [Google Scholar]
- Furukawa K., Arimura N., Miyazaki T. Nucleotide sequence of the 2,3-dihydroxybiphenyl dioxygenase gene of Pseudomonas pseudoalcaligenes. J Bacteriol. 1987 Jan;169(1):427–429. doi: 10.1128/jb.169.1.427-429.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furukawa K., Simon J. R., Chakrabarty A. M. Common induction and regulation of biphenyl, xylene/toluene, and salicylate catabolism in Pseudomonas paucimobilis. J Bacteriol. 1983 Jun;154(3):1356–1362. doi: 10.1128/jb.154.3.1356-1362.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen J. B., Olsen R. H. Isolation of large bacterial plasmids and characterization of the P2 incompatibility group plasmids pMG1 and pMG5. J Bacteriol. 1978 Jul;135(1):227–238. doi: 10.1128/jb.135.1.227-238.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harayama S., Rekik M. Bacterial aromatic ring-cleavage enzymes are classified into two different gene families. J Biol Chem. 1989 Sep 15;264(26):15328–15333. [PubMed] [Google Scholar]
- Kado C. I., Liu S. T. Rapid procedure for detection and isolation of large and small plasmids. J Bacteriol. 1981 Mar;145(3):1365–1373. doi: 10.1128/jb.145.3.1365-1373.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimbara K., Hashimoto T., Fukuda M., Koana T., Takagi M., Oishi M., Yano K. Cloning and sequencing of two tandem genes involved in degradation of 2,3-dihydroxybiphenyl to benzoic acid in the polychlorinated biphenyl-degrading soil bacterium Pseudomonas sp. strain KKS102. J Bacteriol. 1989 May;171(5):2740–2747. doi: 10.1128/jb.171.5.2740-2747.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Merril C. R., Goldman D., Sedman S. A., Ebert M. H. Ultrasensitive stain for proteins in polyacrylamide gels shows regional variation in cerebrospinal fluid proteins. Science. 1981 Mar 27;211(4489):1437–1438. doi: 10.1126/science.6162199. [DOI] [PubMed] [Google Scholar]
- Nakai C., Kagamiyama H., Nozaki M., Nakazawa T., Inouye S., Ebina Y., Nakazawa A. Complete nucleotide sequence of the metapyrocatechase gene on the TOI plasmid of Pseudomonas putida mt-2. J Biol Chem. 1983 Mar 10;258(5):2923–2928. [PubMed] [Google Scholar]
- Nörtemann B., Baumgarten J., Rast H. G., Knackmuss H. J. Bacterial communities degrading amino- and hydroxynaphthalene-2-sulfonates. Appl Environ Microbiol. 1986 Nov;52(5):1195–1202. doi: 10.1128/aem.52.5.1195-1202.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel T. R., Barnsley E. A. Naphthalene metabolism by pseudomonads: purification and properties of 1,2-dihydroxynaphthalene oxygenase. J Bacteriol. 1980 Aug;143(2):668–673. doi: 10.1128/jb.143.2.668-673.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel T. R., Gibson D. T. Purification and propeties of (plus)-cis-naphthalene dihydrodiol dehydrogenase of Pseudomonas putida. J Bacteriol. 1974 Sep;119(3):879–888. doi: 10.1128/jb.119.3.879-888.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sala-Trepat J. M., Evans W. C. The meta cleavage of catechol by Azotobacter species. 4-Oxalocrotonate pathway. Eur J Biochem. 1971 Jun 11;20(3):400–413. doi: 10.1111/j.1432-1033.1971.tb01406.x. [DOI] [PubMed] [Google Scholar]
- Serdar C. M., Gibson D. T. Studies of nucleotide sequence homology between naphthalene-utilizing strains of bacteria. Biochem Biophys Res Commun. 1989 Oct 31;164(2):772–779. doi: 10.1016/0006-291x(89)91526-x. [DOI] [PubMed] [Google Scholar]
- Shamsuzzaman K. M., Barnsley E. A. The regulation of naphthalene metabolism in pseudomonads. Biochem Biophys Res Commun. 1974 Sep 23;60(2):582–589. doi: 10.1016/0006-291x(74)90280-0. [DOI] [PubMed] [Google Scholar]
- Shamsuzzaman K. M., Barnsley E. A. The regulation of naphthalene oxygenase in pseudomonads. J Gen Microbiol. 1974 Jul;83(0):165–170. doi: 10.1099/00221287-83-1-165. [DOI] [PubMed] [Google Scholar]
- Taira K., Hayase N., Arimura N., Yamashita S., Miyazaki T., Furukawa K. Cloning and nucleotide sequence of the 2,3-dihydroxybiphenyl dioxygenase gene from the PCB-degrading strain of Pseudomonas paucimobilis Q1. Biochemistry. 1988 May 31;27(11):3990–3996. doi: 10.1021/bi00411a015. [DOI] [PubMed] [Google Scholar]
- Williams P. A., Catterall F. A., Murray K. Metabolism of naphthalene, 2-methylnaphthalene, salicylate, and benzoate by Pseudomonas PG: regulation of tangential pathways. J Bacteriol. 1975 Nov;124(2):679–685. doi: 10.1128/jb.124.2.679-685.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittich R. M., Rast H. G., Knackmuss H. J. Degradation of naphthalene-2,6- and naphthalene-1,6-disulfonic acid by a Moraxella sp. Appl Environ Microbiol. 1988 Jul;54(7):1842–1847. doi: 10.1128/aem.54.7.1842-1847.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yen K. M., Gunsalus I. C. Plasmid gene organization: naphthalene/salicylate oxidation. Proc Natl Acad Sci U S A. 1982 Feb;79(3):874–878. doi: 10.1073/pnas.79.3.874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zylstra G. J., Gibson D. T. Toluene degradation by Pseudomonas putida F1. Nucleotide sequence of the todC1C2BADE genes and their expression in Escherichia coli. J Biol Chem. 1989 Sep 5;264(25):14940–14946. [PubMed] [Google Scholar]
- Zürrer D., Cook A. M., Leisinger T. Microbial desulfonation of substituted naphthalenesulfonic acids and benzenesulfonic acids. Appl Environ Microbiol. 1987 Jul;53(7):1459–1463. doi: 10.1128/aem.53.7.1459-1463.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]


