Abstract
Fungal elicitor stimulates a multicomponent defense response in cultured parsley cells (Petroselinum crispum). Early elements of this receptor-mediated response are ion fluxes across the plasma membrane and the production of reactive oxygen species (ROS), sequentially followed by defense gene activation and phytoalexin accumulation. Omission of Ca2+ from the culture medium or inhibition of elicitor-stimulated ion fluxes by ion channel blockers prevented the latter three reactions, all of which were triggered in the absence of elicitor by amphotericin B-induced ion fluxes. Inhibition of elicitor-stimulated ROS production using diphenylene iodonium blocked defense gene activation and phytoalexin accumulation. O2− but not H2O2 stimulated phytoalexin accumulation, without inducing proton fluxes. These results demonstrate a causal relationship between early and late reactions of parsley cells to the elicitor and indicate a sequence of signaling events from receptor-mediated activation of ion channels via ROS production and defense gene activation to phytoalexin synthesis. Within this sequence, O2− rather than H2O2 appears to trigger the subsequent reactions.
Keywords: furanocoumarins, ion channels, peptide elicitor, reactive oxygen species, signal transduction
Plants respond to pathogen attack by activating a large variety of defense reactions, including transcriptional activation of genes encoding phenylpropanoid-biosynthetic (1) and various other enzymes and proteins (2, 3), in addition to direct enzyme activation that leads to both reactive oxygen species (ROS) production (“oxidative burst”, ref. 4) and reinforcement of the cell wall (5). Parsley leaf buds respond strongly in this manner when inoculated with spores of Phytophthora sojae, a fungal pathogen of soybean. The overall response comprises formation of small necrotic lesions resulting from hypersensitive cell death, incorporation of phenolic compounds into, and apposition of callose onto, cell walls at the infection site, as well as local and systemic activation of defense-related genes, and secretion of furanocoumarin phytoalexins into the infection droplet (6, 7). With the exception of hypersensitive cell death and callose formation, all of these reactions are also stimulated in cultured parsley cells or protoplasts by treatment either with a crude elicitor preparation from the mycelium of P. sojae (8, 9), with a 42-kDa glycoprotein elicitor from the same fungus (10), or with a 13-amino acid oligopeptide fragment (Pep-13) derived from this protein (11).
Pep-13 has been shown to be necessary and sufficient to stimulate the complete elicitor response in parsley cells, including ion fluxes across the plasma membrane, the oxidative burst, activation of defense-related genes, accumulation of phytoalexins, and production of ethylene (11–13). Binding of radiolabeled Pep-13 to parsley microsomes or protoplasts was found to be specific, reversible, and saturable (11). A 91-kDa plasma membrane protein was specifically labeled by covalent crosslinking of Pep-13 to parsley membranes (14). Because identical structural elements of Pep-13 were required for specific binding, crosslinking and activation of defense reactions (11, 14), this membrane protein appears to represent the elicitor receptor through which the entire defense response is initiated.
In parsley as well as other plant cells, pathogen defense reactions are induced by elicitor only if Ca2+ is present in the culture medium (11, 15–17). In tobacco, soybean, carrot, and parsley, this requirement correlates with rapid elicitor-stimulated Ca2+ influx (11, 18–20), suggesting that Ca2+ is an important element in elicitor-mediated signal transduction, as it is in many other signaling processes (21, 22). This and other early responses to elicitor treatment, such as various ion fluxes across the plasma membrane, synthesis of ROS, and phosphorylation and dephosphorylation of proteins, have frequently been discussed as putative components of signal transduction chain(s) leading to defense gene activation and/or hypersensitive cell death (4, 15–17, 23–26). However, unequivocal evidence for a causal relationship and a sequential order of the signaling elements is still lacking. Here we report that the elicitor-stimulated oxidative burst in cultured parsley cells depends on ion fluxes across the plasma membrane and is necessary and sufficient for triggering phytoalexin accumulation.
MATERIALS AND METHODS
Cell Culture and Elicitor Treatment.
Cell suspension cultures of Petroselinum crispum were propagated according to a published procedure (27). Protoplast preparation 5 days after subculturing, treatment with elicitor for 24 hr, and quantification of furanocoumarins were performed as described (8). Cell viability was determined by double staining with fluorescein diacetate and propidium iodide (28). Crude cell-wall elicitor was prepared from the mycelium of Phytophthora sojae, race 1 (27). The oligopeptide elicitor, Pep-13 (11), was synthesized by Kem-En-Tec (Copenhagen). The polyene antibiotics, amphotericin B and nystatin, diphenylene iodonium (DPI), the ion channel blockers, and KO2 were applied in dimethyl sulfoxide [final solvent concentration, 0.1% (vol/vol)].
Measurement of Ion Fluxes and ROS.
Ion concentrations were determined using ion-selective electrodes for H+, K+, and Cl− or by monitoring the uptake of 45Ca2+ (11). H2O2 release into the culture medium was quantified by measuring chemiluminescence produced by ferricyanide-catalyzed luminol oxidation (29). Briefly, 50 μl medium were added to 750 μl phosphate buffer (50 mM potassium phosphate, pH 7.9) prior to automated injection of 200 μl luminol (0.3 mM in phosphate buffer) and 100 μl K3[Fe(CN)6] (14 mM in H2O) by the luminometer, Lumat LB 9501 (Berthold, Wildbach, Germany). Chemiluminescence was recorded 3 sec after the last injection with a signal integration time of 5 sec.
For O2− determination (30), 0.5 mM sodium diethyldithiocarbamate (DDC) was added to the cell suspension culture to inhibit superoxide dismutases. At different times after addition of elicitor, 50-μl aliquots were removed from the culture medium and added to 750 μl glycine-NaOH buffer (100 mM glycine, pH 9.0/1 mM EDTA), immediately followed by automatic injection of 200 μl lucigenin solution (550 μM lucigenin in glycine-NaOH buffer) and 100 μl glycine-NaOH buffer. Chemiluminescence was recorded for 10 sec after the last injection. The signal was calibrated using 50 μM xanthine and 0.025–25 milliunits xanthine oxidase or KO2.
RNA Isolation, Blot Hybridization, and Run-On Transcription.
Total RNA from parsley protoplasts was prepared as described (8) and heat-denatured at 65°C for 15 min. RNA samples (7.5 μg) were electrophoresed in agarose using 3-(N-morpholino)propanesulfonic acid-EDTA buffer and transferred to nylon filters. UV crosslinking of RNA to the filter, prehybridization, hybridization, and 32P-labeling of cDNA probes were carried out as described (31). Transcriptionally active nuclei were isolated from cultured parsley cells (32, 33) and used in run-on transcription experiments (9, 33). Parsley cDNA probes encoding 18 elicitor-responsive genes (9, 34), one light-responsive gene (33), and two constitutively expressed genes (9, 34) were employed in these analyses.
RESULTS
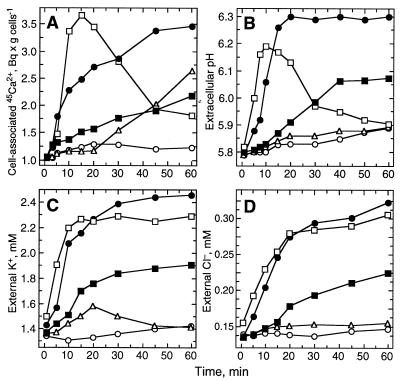
Elicitor treatment stimulated rapid and transient uptake of 45Ca2+ by cultured parsley cells (Fig. 1A). The increase in Ca2+ flux commenced within 2–4 min, thus representing one of the earliest detectable reactions of the cells. Increases in extracellular Cl− and K+ concentrations and alkalinization of the culture medium were stimulated with similar kinetics, indicating concomitant effluxes of Cl− and K+ and uptake of protons (Fig. 1 B–D). Because all of these elicitor-stimulated ion fluxes occurred along their concentration gradients, various ionophores were tested for their ability to simulate these fluxes and thereby activate defense reactions in the absence of elicitor. Nigericin, A23187, valinomycin, and ionomycin stimulated Ca2+ and K+ fluxes, but only nigericin and A23187 stimulated Cl− fluxes as well. However, all of these compounds failed to stimulate alkalinization of the culture medium or phytoalexin accumulation in the absence of elicitor. The saponin, digitonin, which induces callose but not phytoalexin synthesis (35), stimulated Ca2+ influx (Fig. 1A) but none of the other ion fluxes (Fig. 1 B–D). The polyene antibiotics, amphotericin B and nystatin, stimulated ion fluxes qualitatively similar to those observed in response to elicitor; however, the relative intensities and durations of the individual fluxes varied greatly (Fig. 1). The changes in ion concentrations caused by amphotericin B reached ≈50% of the elicitor-stimulated levels, though at considerably lower initial rates, and were of similar duration. In contrast, nystatin induced external Cl− and K+ levels almost identical to those induced by elicitor, and very similar initial rates of Ca2+ uptake and medium alkalinization. However, the changes in intracellular Ca2+ levels and medium alkalinization occurred only transiently. Nystatin failed to induce phytoalexin accumulation in parsley cells and protoplasts, in contrast to amphotericin B and elicitor (35), both of which induced the formation of the same relative amounts of the various furanocoumarin derivatives with similar efficiency.
Figure 1.
Time courses of changes in Ca2+ (A), H+ (B), K+ (C) and Cl− (D) concentrations stimulated in suspension-cultured parsley cells by treatment with crude cell-wall elicitor (50 μg/ml; •), amphotericin B (50 μM, ▪), nystatin (50 μM, □), digitonin (50 μM, ▵), or sterile water (○).
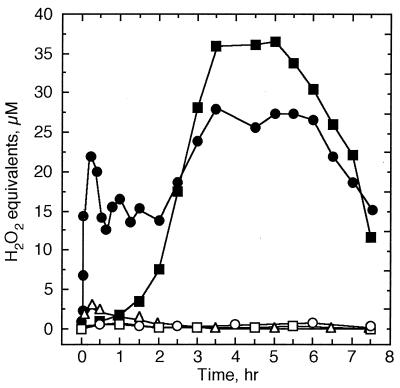
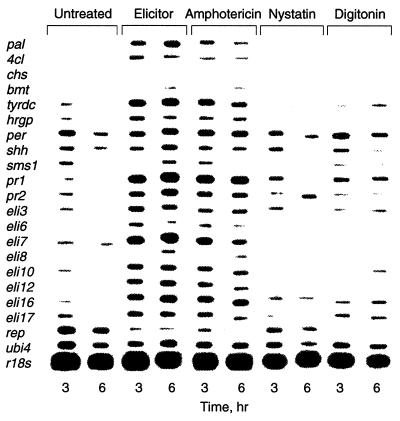
Another almost immediate reaction of parsley cells to elicitor treatment was the oxidative burst, measured as an increase in luminol-mediated chemiluminescence caused by H2O2 in the culture medium. Within 4–6 min after addition of the elicitor, the H2O2 concentration of the culture medium began to increase, reaching a first maximum of ≈20 μM at 15–20 min and a second, more sustained one at 3–6 hr (Fig. 2). Maximal concentrations (≈25–30 μM) reflected about a 50-fold increase above the level in the medium of untreated cells (<0.6 μM H2O2). Similar timing and extent of O2− production were observed with lucigenin in the medium of elicitor-treated cells, when DDC was used to inhibit superoxide dismutases. In the absence of elicitor, DDC treatment of parsley cells resulted in a steady O2− accumulation up to ≈70% of the oxidative burst maximum. In the absence of DDC, O2− was not detectable. These results suggested that the O2−-generating system was continuously active at low rates, producing ROS that were probably removed immediately by extracellular superoxide dismutases or other protective systems. No reduction in cell viability was observed within 24 hr of elicitor treatment. Amphotericin B caused a massive but comparatively slow release of H2O2 into the culture medium, whereas the structurally related polyene antibiotic, nystatin, and digitonin were incapable of stimulating this response (Fig. 2). The ionophores, A23187, ionomycin, nigericin, and valinomycin, also failed to stimulate an oxidative burst. Analogous results were obtained at the level of defense gene activation. Results from run-on transcription of 18 defense-related genes as well as one elicitor-repressed (rep), one light-responsive (chs), and two constitutively expressed genes (ubi4 and r18s) are shown in Fig. 3. While amphotericin B activated all elicitor-responsive genes, nystatin and digitonin induced the transcription of distinct subsets of these genes.
Figure 2.
Time courses of H2O2 accumulation in the medium of cultured parsley cells treated with crude cell wall-elicitor (50 μg/ml; •), amphotericin B (50 μM, ▪), nystatin (50 μM, □), digitonin (50 μM, ▵), or water (○).
Figure 3.
Run-on transcription of defense-related genes in nuclei isolated from cultured parsley cells 3 or 6 hr after treatment with water (“untreated”), crude cell-wall elicitor (50 μg/ml), amphotericin B (50 μM), nystatin (50 μM), or digitonin (50 μM). The homologous cDNA probes used for hybridization encode phenylalanine ammonia-lyase (pal), 4-coumarate:CoA ligase (4cl), chalcone synthase (chs), S-adenosyl-l-methionine:bergaptol O-methyltransferase (bmt), tyrosine decarboxylase (tyrdc), hydroxyproline-rich glycoprotein (hrgp), anionic peroxidase (per), S-adenosyl-l-homocystein hydrolase (shh), S-adenosyl-l-methionine synthetase 1 (sms1), intracellular pathogenesis-related proteins 1 and 2 (pr1 and pr2), proteins of unknown function (eli3, eli6, eli7, eli8, eli10, eli12, eli16, eli17, rep), polyubiquitin (ubi4), and ribosomal 18S RNA (r18s).
Several ion channel blockers, belonging to the classes of anthracene-9-carboxylate (A-9-C), diphenylamine-2-carboxylate and indanyloxyacetate (IAA-94) derivatives, were tested with respect to their effects on elicitor-inducible reactions. The same inhibitors have been applied successfully in both mammalian (36) and plant tissues (37). As shown in Table 1, all ion channel blockers analyzed inhibited elicitor-induced Cl− and K+ effluxes, H2O2 production, and phytoalexin accumulation. Similar inhibitor concentrations were required to cause a 50% inhibition of these reactions. The A-9-C derivatives were slightly less effective in inhibiting medium alkalinization, and the diphenylamine-2-carboxylates and IAA-94 did not affect elicitor-stimulated Ca2+ influx at all, suggesting that the different classes of ion channel blockers acted on separate target sites. Benzoic acid (up to 500 μM), which is frequently used as an inactive analog of A-9-C in electrophysiological experiments, inhibited neither the oxidative burst nor furanocoumarin synthesis. The same was true for 5-nitro-2-[3-(4-aminophenyl)propylamino]benzoic acid (NH2-NPPB), an inactive amino derivative of the Cl− channel blocker 5-nitro-2-(3-phenylpropylamino)benzoic acid (NPPB) (38). The viability of parsley protoplasts remained unchanged within 24 hr of treatment with inhibitory concentrations of any of the ion channel blockers tested. Benzoic acid, A-9-C, niflumic acid, and IAA-94 did not affect the mRNA level of the constitutively expressed gene, ubi4, whereas elicitor-induced pal, 4cl, tyrdc, and eli12 mRNA accumulation was strongly inhibited by 10 μM A-9-C, 20 μM niflumic acid, or 100 μM IAA-94, but not by 10 μM benzoic acid.
Table 1.
Inhibition of elicitor-stimulated reactions in cultured parsley cells by anion channel blockers
| Inhibitor | IC50, μM
|
|||||
|---|---|---|---|---|---|---|
| Ca2+ influx | K+ efflux | Cl− efflux | H+ influx | H2O2 | Phytoalexins | |
| Anthracene-9-carboxylate | ||||||
| Anthracene-9-carboxylate | 15 | 24 | 16 | 87 | 39 | 4 |
| Anthracene-9-carbaldehyde | 7 | 40 | 55 | 87 | 20 | 10 |
| Anthraquinone-2-carboxylate | 15 | 17 | 12 | 31 | 15 | 3 |
| Xanthene-9-carboxylate | ND | 30 | 16 | 108 | 34 | 8 |
| 2-Aminoanthracene* | ND | — | — | — | — | — |
| Benzoic acid* | — | — | — | — | — | — |
| Diphenylamine-2-carboxylate | ||||||
| Niflumic acid | — | 3 | 7 | 19 | 15 | 11 |
| Flufenamic acid | — | 11 | 1 | 17 | 2 | 11 |
| Mefenamic acid | — | 11 | 4 | 78 | 6 | 12 |
| Tolfenamic acid | — | 3 | 3 | 33 | 11 | 5 |
| NPPB | — | 28 | 5 | 36 | 35 | 10 |
| NH2-NPPB* | ND | ND | ND | — | — | — |
| IAA-94 | — | 86 | 160 | 300 | 150 | 47 |
Concentrations required to inhibit the individual reactions by 50% (IC50) were determined by treating cells with the inhibitor 15 min before addition of crude cell wall elicitor (50 μg/ml) and measuring the indicated reactions 30 min (Ca2+, K+, Cl−, H+, H2O2) or 24 hr (phytoalexins) after initiation of elicitor treatment {ND, not determined; —, no effect; NPPB, 5-nitro-2-(3-phenylpropylamino)benzoate; NH2-NPPB, 5-nitro-2-[3-(4-aminophenyl)proylamino]benzoate; IAA-94, indanyloxyacetate 94; ∗, inactive derivative}.
To evaluate the role of H2O2 in elicitor signal transduction, parsley cells were treated with catalase (2 mkat/l) 10 min before the addition of elicitor. Catalase completely abolished detection of H2O2 by the luminol assay, indicating that luminol oxidation indeed was caused by H2O2 from the oxidative burst. However, catalase treatment did not affect elicitor-induced phytoalexin synthesis, nor did external application of H2O2 (0.1 μM to 20 mM) or generation of H2O2 in the culture medium by addition of 0.3–670 units/liter glucose oxidase and 5 mM glucose stimulate phytoalexin accumulation. On the other hand, inhibition of the oxidative burst by DPI, a suicide substrate inhibitor of the mammalian NADPH oxidase (39), blocked phytoalexin formation without affecting cell viability and elicitor-stimulated alkalinization of the culture medium. The IC50 concentrations required to half-maximally inhibit H2O2 and phytoalexin accumulation were 0.8–1.2 and 1.0–2.1 μM DPI, respectively, similar to the IC50 value of 0.5 μM reported for the macrophage NADPH oxidase (40). The corresponding IC50 values for diphenyl iodonium, a less potent inhibitor of the mammalian NADPH oxidase, were 152–178 and 115–180 μM, respectively, as compared with 80 μM for inhibition of the macrophage NADPH oxidase (40). A concentration of 5 μM DPI was also sufficient to strongly inhibit the elicitor-induced accumulation of various defense-related mRNAs without affecting the ubi4 mRNA level.
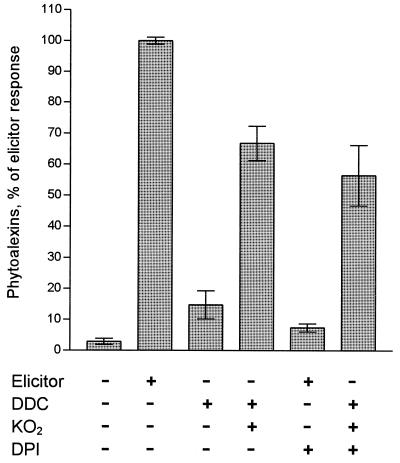
These results suggested that the primary product of the NAD(P)H oxidase reaction, O2−, was involved in transmitting the elicitor signal, even though O2− accumulation was detectable in the culture medium only in the presence of the superoxide dismutase inhibitor, DDC. The finding that elicitor treatment of parsley cells in the presence of increasing amounts of superoxide dismutase (1–35 μkat/ml) at pH 7 inhibited phytoalexin production up to 60% in a dose-dependent manner further supported this conclusion. Addition of DDC to parsley cells in the absence of elicitor stimulated phytoalexin accumulation to 18 ± 4% of the elicitor response, suggesting that the amount of O2− generated when its rapid degradation was inhibited by DDC were sufficient to trigger this reaction. Because xanthine oxidase alone greatly inhibited all elicitor-stimulated reactions, direct gain-of-function experiments were performed by adding KO2 to the culture medium. In the presence of DDC, KO2 stimulated the production of phytoalexins in the absence of elicitor to levels of 67 ± 6% of the elicitor response (Fig. 4), whereas it was inactive in the absence of DDC. In a control experiment, addition of equivalent amounts of KCl to cultured parsley cells in the presence of DDC did not stimulate phytoalexin accumulation. The furanocoumarins induced by KO2 were the same as those observed after elicitor treatment. KO2 treatment also abolished the inhibitory effect of DPI on elicitor-induced phytoalexin synthesis, in contrast to H2O2 which had no effect.
Figure 4.
Induction of phytoalexin production in cultured parsley cells by O2−. KO2 (250 μM, suspended in dimethyl sulfoxide [0.1%]) was added to the culture medium. Equivalent amounts of dimethyl sulfoxide were added to the incubations without KO2. For comparison, parsley cells were treated with the peptide elicitor, Pep-13 (400 nM). The inhibitors DDC and DPI were applied at 0.5 mM and 10 μM, respectively. Error bars indicate standard deviations (9–28 independent experiments) within a 95% confidence level.
DISCUSSION
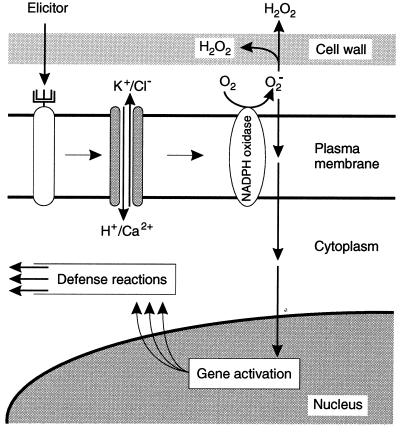
Our present results indicate the causal connections and the sequence of elicitor-induced reactions depicted in Fig. 5. Ion fluxes across the plasma membrane were prerequisites for all subsequent reactions in cultured parsley cells, thus representing the most upstream-located elements so far identified within the signal cascade. Patch-clamp analysis of parsley protoplasts recently identified one particular ion channel that was transiently activated by elicitor to mediate Ca2+ influx under physiological conditions (41). However, the existence of additional, directly or indirectly elicitor-responsive ion channels in the plasma membrane must be postulated, since Ca2+ fluxes are obviously necessary but not sufficient for the initiation of downstream reactions. This hypothesis gains further support from the different potencies of ion channel inhibitors to block the individual elicitor-stimulated ion fluxes. Moreover, the involvement of distinct types of Ca2+ channels in elicitor signal transduction in different plants was suggested by the finding that piperazines inhibit Ca2+ fluxes and phytoalexin accumulation in parsley (17) but not in soybean cells (18). In contrast, La3+ efficiently inhibited both elicitor-stimulated reactions in parsley (41) as well as soybean (42, 43), carrot (44), and tobacco (19).
Figure 5.
Hypothetical scheme for elicitor-induced signal transduction in parsley.
Major differences among species were also observed in gain-of-function experiments. While similar durations and intensities of all four elicitor-stimulated ion fluxes appear to be required to activate the oxidative burst, defense gene transcription and phytoalexin synthesis in parsley in the absence of elicitor, the Ca2+ ionophore A23187 alone stimulated phytoalexin accumulation in soybean (42) and carrot cells (44). In tobacco cells, A23187 mimicked elicitor-stimulated Ca2+ fluxes and medium alkalinization but did not induce phytoalexin production (19). A common prerequisite in all of these experimental systems is the elicitor-stimulated activation of Ca2+ influx for initiation of all subsequent defense reactions. In some systems, these Ca2+ fluxes across the plasma membrane are apparently sufficient, whereas in others additional ion channels and pumps seem to be required for the induction of downstream effects. Plasma membrane-located ion channels have also been identified as important components of signal transduction cascades in plant responses to various other environmental and hormonal signals (45). In this context it is interesting to note that different sets of genes are transcriptionally activated in parsley cells upon variation of the nature, duration, and intensity of ion fluxes, possibly indicating a simple means of specifically converting many different external signals into appropriate gene expression patterns by modulating the activities of a comparatively small number of ion channels.
We have now shown that, in parsley cells, elicitor-stimulated ROS production was a prerequisite for defense gene activation but not for the stimulation of ion fluxes, and that inhibitors of elicitor-stimulated ion fluxes blocked the oxidative burst as well as defense gene activation and phytoalexin accumulation. Thus, the oxidative burst must be localized downstream of the ion flux changes, but upstream of defense gene activation, which in turn precedes phytoalexin production. Furthermore, the loss and gain-of-function experiments demonstrate that O2− (or a product derived from O2−) rather than H2O2 is an essential element of the signal cascade leading to phytoalexin production. O2− generated by a plasma membrane-located NAD(P)H oxidase was initially suggested by Doke et al. (46) to be involved in triggering phytoalexin synthesis and hypersensitive cell death in potato. Different experimental systems have since been employed to investigate possible causal links between ROS and these defense reactions, but have not yielded unambiguous results (4, 25). In contrast to the results presented here for parsley, DPI was reported not to affect elicitor-induced chalcone synthase mRNA accumulation in soybean (47), nor phytoalexin production in tobacco cells (48), although the oxidative burst was inhibited in both cases. In soybean cells, both loss and gain-of-function experiments indicated that the oxidative burst was involved in triggering hypersensitive cell death (47, 49). Strong correlations between H2O2 production and cell death were also observed in incompatible interactions of cultured tobacco and soybean cells with Pseudomonas syringae (25, 50). This correlation was lost, however, when tobacco cells were cocultivated with a P. syringae strain carrying the avr gene together with mutations in the hrmA region (50). In this interaction, the plant cells responded with an oxidative burst but showed no hypersensitive cell death, suggesting that ROS from the oxidative burst were necessary but not sufficient for triggering cell death. This interpretation gains further support from our finding that elicitor treatment of parsley cells did not induce cell death, although the oxidative burst caused by this treatment was very similar to that observed in soybean and tobacco cells cocultivated with avirulent bacterial strains (25, 50). Furthermore, O2− rather than H2O2 was demonstrated to be necessary and sufficient for induction of lesion formation and PR-1 mRNA accumulation in the “lesion simulating disease resistance response” mutant, lsd1, of Arabidopsis thaliana (51). Thus, accumulating evidence supports the notion that ROS from the oxidative burst are involved in pathogen defense-related signaling in plants. It remains to be established, whether contrasting results obtained with different systems regarding the nature of individual ROS species that mediate defense gene activation and/or cell death reflect differences in experimental detail or species specificity of the signaling mechanisms.
Acknowledgments
The technical assistance of K. Adamitza, H. Nixdorf, and D. Reinhardt, the secretarial assistance of E. Deffner and R. Laue, and the art work of C. Kaufmann are gratefully acknowledged. We thank Drs. T. Nürnberger (Institut für Pflanzenbiochemie, Halle, Germany) and R. Cormack (Max-Planck-Institut, Köln, Germany) for valuable discussions, and Prof. R. Greger (Universität Freiburg, Germany) for providing 5-nitro-2-[3-(4-aminophenyl)propylamino]benzoic acid. This work was supported by the Deutsche Forschungsgemeinschaft (Sche 235/3–1, 3–2 and 3–3), the European Community (ERBCHRXCT 930168), and the Fonds der Chemischen Industrie.
ABBREVIATIONS
- DDC
sodium diethyldithiocarbamate
- DPI
diphenylene iodonium
- Pep-13
oligopeptide elicitor
- ROS
reactive oxygen species
References
- 1.Hahlbrock K, Scheel D. Annu Rev Plant Physiol Plant Mol Biol. 1989;40:347–369. [Google Scholar]
- 2.Bowles D J. Annu Rev Biochem. 1990;59:873–907. doi: 10.1146/annurev.bi.59.070190.004301. [DOI] [PubMed] [Google Scholar]
- 3.Boller T. In: Plant-Microbe Interactions. Kosuge T, Nester E W, editors. Vol. 2. New York: Macmillan; 1987. pp. 385–413. [Google Scholar]
- 4.Mehdy M C, Sharma Y K, Sathasivan K, Bays N W. Physiol Plant. 1996;98:365–374. [Google Scholar]
- 5.Bradley D J, Kjellbom P, Lamb C J. Cell. 1992;70:21–30. doi: 10.1016/0092-8674(92)90530-p. [DOI] [PubMed] [Google Scholar]
- 6.Scheel D, Hauffe K D, Jahnen W, Hahlbrock K. In: Recognition in Microbe-Plant Symbiotic and Pathogenic Interactions. Lugtenberg B J J, editor. H4. Berlin: Springer; 1986. pp. 325–331. [Google Scholar]
- 7.Schmelzer E, Krüger-Lebus S, Hahlbrock K. Plant Cell. 1989;1:993–1001. doi: 10.1105/tpc.1.10.993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dangl J L, Hauffe K D, Lipphardt S, Hahlbrock K, Scheel D. EMBO J. 1987;6:2551–2556. doi: 10.1002/j.1460-2075.1987.tb02543.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Somssich I E, Bollmann J, Hahlbrock K, Kombrink E, Schulz W. Plant Mol Biol. 1989;12:227–234. doi: 10.1007/BF00020507. [DOI] [PubMed] [Google Scholar]
- 10.Parker J E, Schulte W, Hahlbrock K, Scheel D. Mol Plant-Microbe Interact. 1991;4:19–27. [Google Scholar]
- 11.Nürnberger T, Nennstiel D, Jabs T, Sacks W R, Hahlbrock K, Scheel D. Cell. 1994;78:449–460. doi: 10.1016/0092-8674(94)90423-5. [DOI] [PubMed] [Google Scholar]
- 12.Sacks W R, Nürnberger T, Hahlbrock K, Scheel D. Mol Gen Genet. 1995;246:45–55. doi: 10.1007/BF00290132. [DOI] [PubMed] [Google Scholar]
- 13.Hahlbrock K, Scheel D, Logemann E, Nürnberger T, Parniske M, Reinold S, Sacks W R, Schmelzer E. Proc Natl Acad Sci USA. 1995;92:4150–4157. doi: 10.1073/pnas.92.10.4150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nürnberger T, Nennstiel D, Hahlbrock K, Scheel D. Proc Natl Acad Sci USA. 1995;92:2338–2342. doi: 10.1073/pnas.92.6.2338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dietrich A, Mayer J E, Hahlbrock K. J Biol Chem. 1990;265:6360–6368. [PubMed] [Google Scholar]
- 16.Ebel J, Scheel D. In: Genes Involved in Plant Defense. Boller T, Meins F, editors. Vienna: Springer; 1992. pp. 183–205. [Google Scholar]
- 17.Nürnberger T, Colling C, Hahlbrock K, Jabs T, Renelt A, Sacks W R, Scheel D. Biochem Soc Symp. 1994;60:173–182. [PubMed] [Google Scholar]
- 18.Ebel J, Bhagwat A A, Cosio E G, Feger M, Kissel U, Mithöfer A, Waldmüller T. Can J Bot. 1995;73:5506–5510. [Google Scholar]
- 19.Tavernier E, Wendehenne D, Blein J-P, Pugin A. Plant Physiol. 1995;109:1025–1031. doi: 10.1104/pp.109.3.1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bach M, Schnitzler J-P, Seitz H U. Plant Physiol. 1993;103:407–412. doi: 10.1104/pp.103.2.407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bush D S. Plant Physiol. 1993;103:7–13. doi: 10.1104/pp.103.1.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Clapham D E. Cell. 1995;80:259–268. doi: 10.1016/0092-8674(95)90408-5. [DOI] [PubMed] [Google Scholar]
- 23.Dixon R A, Lamb C J. Annu Rev Plant Physiol Plant Mol Biol. 1990;41:339–367. doi: 10.1146/annurev.arplant.48.1.251. [DOI] [PubMed] [Google Scholar]
- 24.Boller T. Annu Rev Plant Physiol Plant Mol Biol. 1995;46:189–214. [Google Scholar]
- 25.Baker C J, Orlandi E W. Annu Rev Phytopathol. 1995;33:299–321. doi: 10.1146/annurev.py.33.090195.001503. [DOI] [PubMed] [Google Scholar]
- 26.Jones A M, Dangl J L. Trends Plant Sci. 1996;1:114–119. [Google Scholar]
- 27.Kombrink E, Hahlbrock K. Plant Physiol. 1986;81:216–221. doi: 10.1104/pp.81.1.216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huang C-N, Cornejo M J, Bush D S, Jones R L. Protoplasma. 1986;135:80–87. [Google Scholar]
- 29.Schwacke R, Hager A. Planta. 1992;187:136–141. doi: 10.1007/BF00201635. [DOI] [PubMed] [Google Scholar]
- 30.Corbisier P, Houbion A, Remacle J. Anal Biochem. 1987;164:240–247. doi: 10.1016/0003-2697(87)90392-7. [DOI] [PubMed] [Google Scholar]
- 31.Kawalleck P, Keller H, Hahlbrock K, Scheel D, Somssich I E. J Biol Chem. 1993;268:2189–2194. [PubMed] [Google Scholar]
- 32.Willmitzer L, Wagner K G. Exp Cell Res. 1981;135:69–77. doi: 10.1016/0014-4827(81)90300-1. [DOI] [PubMed] [Google Scholar]
- 33.Chappell J, Hahlbrock K. Nature (London) 1984;311:76–78. [Google Scholar]
- 34.Van Gijsegem F, Somssich I E, Scheel D. Eur J Plant Pathol. 1995;101:549–559. [Google Scholar]
- 35.Scheel D, Colling C, Hedrich R, Kawalleck P, Parker J E, Sacks W R, Somssich I E, Hahlbrock K. In: Advances in Molecular Genetics of Plant-Microbe Interactions. Hennecke H, Verma D P S, editors. Vol. 1. Dordrecht, The Netherlands: Kluwer; 1991. pp. 373–380. [Google Scholar]
- 36.Gögelein H. Biochim Biophys Acta. 1988;947:521–547. doi: 10.1016/0304-4157(88)90006-8. [DOI] [PubMed] [Google Scholar]
- 37.Marten I, Zeilinger C, Redhead C, Landry D W, Al-Awqati Q, Hedrich R. EMBO J. 1992;11:3569–3575. doi: 10.1002/j.1460-2075.1992.tb05440.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wangemann P, Wittner M, Di Stefano A, Englert H C, Lang H J, Schlatter E, Greger R. Pflügers Arch Eur J Physiol. 1986;407:S128–S141. doi: 10.1007/BF00584942. [DOI] [PubMed] [Google Scholar]
- 39.O’Donnell V B, Tew D G, Jones O T G, England P J. Biochem J. 1993;290:41–49. doi: 10.1042/bj2900041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hancock J T, Jones O T G. Biochem J. 1987;242:103–107. doi: 10.1042/bj2420103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zimmermann, S., Nürnberger, T., Frachisse, J.-M., Wirtz, W., Guern, J., Hedrich, R. & Scheel, D. (1997) Proc. Natl. Acad. Sci. USA, in press. [DOI] [PMC free article] [PubMed]
- 42.Stäb M R, Ebel J. Arch Biochem Biophys. 1987;257:416–423. doi: 10.1016/0003-9861(87)90585-6. [DOI] [PubMed] [Google Scholar]
- 43.Atkinson M M, Midland S L, Sims J J, Keen N T. Plant Physiol. 1996;112:297–302. doi: 10.1104/pp.112.1.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kurosaki F, Tsurusawa Y, Nishi A. Phytochemistry. 1987;26:1919–1923. [Google Scholar]
- 45.Ward J M, Pel Z-M, Schroeder J I. Plant Cell. 1995;7:833–844. doi: 10.1105/tpc.7.7.833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Doke N, Chai H B, Kawaguchi A. In: Molecular Determinants of Plant Diseases. Nishimura S, Vance C P, Doke N, editors. Tokyo/Springer, Berlin: Japan Sci. Soc. Press; 1987. pp. 235–251. [Google Scholar]
- 47.Levine A, Tenhaken R, Dixon R, Lamb C. Cell. 1994;79:583–593. doi: 10.1016/0092-8674(94)90544-4. [DOI] [PubMed] [Google Scholar]
- 48.Rustérucci C, Stallaert V, Milat M-L, Pugin A, Ricci P, Blein J-P. Plant Physiol. 1996;111:885–891. doi: 10.1104/pp.111.3.885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Levine A, Pennell R I, Alvarez M E, Palmer R, Lamb C. Curr Biol. 1996;6:427–437. doi: 10.1016/s0960-9822(02)00510-9. [DOI] [PubMed] [Google Scholar]
- 50.Glazener J A, Orlandi E W, Baker C J. Plant Physiol. 1996;110:759–763. doi: 10.1104/pp.110.3.759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jabs T, Dietrich R A D, Dangl J L. Science. 1996;273:1853–1856. doi: 10.1126/science.273.5283.1853. [DOI] [PubMed] [Google Scholar]