Abstract
The Sm and Sm-like proteins are conserved in all three domains of life and have emerged as important players in many different RNA-processing reactions. Their proposed role is to mediate RNA–RNA and/or RNA–protein interactions. In marked contrast to eukaryotes, bacteria appear to contain only one distinct Sm-like protein belonging to the Hfq family of proteins. Similarly, there are generally only one or two subtypes of Sm-related proteins in archaea, but at least one archaeon, Methanococcus jannaschii, encodes a protein that is related to Hfq. This archaeon does not contain any gene encoding a conventional archaeal Sm-type protein, suggesting that Hfq proteins and archaeal Sm-homologs can complement each other functionally. Here, we report the functional characterization of M. jannaschii Hfq and its crystal structure at 2.5 Å resolution. The protein forms a hexameric ring. The monomer fold, as well as the overall structure of the complex is similar to that found for the bacterial Hfq proteins. However, clear differences are seen in the charge distribution on the distal face of the ring, which is unusually negative in M. jannaschii Hfq. Moreover, owing to a very short N-terminal α-helix, the overall diameter of the archaeal Hfq hexamer is significantly smaller than its bacterial counterparts. Functional analysis reveals that Escherichia coli and M. jannaschii Hfqs display very similar biochemical and biological properties. It thus appears that the archaeal and bacterial Hfq proteins are largely functionally interchangeable.
Keywords: Hfq, Methanococcus jannaschii, RNA-binding protein, Sm, sRNA, translational regulation
INTRODUCTION
Sm and Sm-like (Lsm) proteins represent a large family of structurally and functionally related proteins of ancient origin. The proteins are ubiquitous in eukaryotes (>18 paralogs) and have the capacity to form at least six different hetero-heptameric ring-shaped complexes that participate in numerous RNA-processing reactions through their association with U-rich target RNAs (for review, see Wilusz and Wilusz 2005). The particular combination of the eukaryotic Sm/Lsm proteins present in a given complex determines its localization as well as the RNA specificity of the complex. Archaeal genomes usually encode either a single or a few small Sm-related proteins. These form homo-hexameric or homo-heptameric rings and share oligomerization behavior and RNA-binding properties with their eukaryotic counterparts (for reviews, see Pannone and Wolin 2000; Beggs 2005; Khusial et al. 2005; Wilusz and Wilusz 2005). The function of the archaeal proteins is currently not clear. However, the Archaeoglobus fulgidus Sm1- and Sm2-type proteins have been shown to associate with RNase P, suggesting a role in post-transcriptional processing of rRNA and tRNA (Törö et al. 2001).
More recently, it has been shown that bacterial Hfq proteins are homologs of the eukaryotic Lsm proteins (Møller et al. 2002a; Zhang et al. 2002). Hfq monomers assemble to form highly stable hexamers, which have a binding preference for A/U-rich tracks on the target RNA (Senear and Steitz 1976; de Haseth and Uhlenbeck 1980). The protein has emerged as a pleitropic post-transcriptional regulator that modulates the turnover and/or the translation of a large number of mRNAs (Muffler et al. 1997; Nogueira and Springer 2000). In line with this, Hfq has been shown to interact with numerous small trans-encoded regulatory RNAs (sRNA) that act by base-pairing with target mRNAs. Hfq has been shown to be absolutely required for the proper function of this class of ribo-regulators (for review, see Gottesman 2004; Storz et al. 2004). One role of Hfq in this context is to protect the sRNAs against degradation. In addition, Hfq strongly promotes the intermolecular base pairing of sRNAs to their target mRNAs, thereby enabling regulation of translation and mRNA stability (Møller et al. 2002a; Zhang et al. 2002; Kawamoto et al. 2006). Thus, the bacterial Hfq system possesses striking similarities in both structure and function to the eukaryotic Lsm proteins (for reviews, see Valentin-Hansen et al. 2004; Wilusz and Wilusz 2005).
Members of the Sm/Lsm family possess a common bipartite motif (∼70 amino acid residues) consisting of two relatively conserved regions termed the Sm1 and Sm2 motifs. These motifs are separated by a region, which is not conserved in its sequence or length, the variable region (Cooper et al. 1995; Hermann et al. 1995; Séraphin 1995). Crystal structures of several eukaryotic and archaeal Sm/Lsm proteins have established that the conserved signature sequence defines a specific Sm-fold composed of a strongly bent five-stranded, antiparallel β-sheet capped by an N-terminal α-helix (Kambach et al. 1999; Collins et al. 2001; Mura et al. 2001, 2003; Törö et al. 2001, 2002; Thore et al. 2003; Kilic et al. 2005). Though bacterial Hfq proteins only exhibit faint sequence similarity to the Sm family of proteins, crystal structures of the Staphylococcus aureus, Escherichia coli, and Pseudomonas aeruginosa homologs (Sau-, Eco-, and Pae-Hfq) have revealed that the protein does indeed contain the distinctive Sm fold (Schumacher et al. 2002; Sauter et al. 2003; Nikulin et al. 2005). Moreover, insight into the RNA-binding mechanism of both the archaeal Sm-like proteins and bacterial Hfq has been gained by the crystal structures of the proteins in complex with small RNA oligonucleotides (Törö et al. 2001; Schumacher et al. 2002; Thore et al. 2003).
Hfq is conserved among a wide range of bacteria (Sun et al. 2002), however, its prevalence is not restricted to the bacterial domain. Database searches identified a potential homolog in the thermophilic archaeon, Methanococcus jannaschii, which does not contain any Sm/Lsm gene (Sauter et al. 2003; Valentin-Hansen et al. 2004). Alignment of the M. jannaschii protein sequence (Mja-Hfq) with the Hfq proteins from S. aureus and E. coli indicates that Mja-Hfq consists of an isolated Sm domain without the flanking C-terminal region found in many of the bacterial proteins (Fig. 1). In E. coli, this C-terminal domain of ∼30 amino acids, can be removed without affecting the activity of the protein (Tsui et al. 1994; Sonnleitner et al. 2002). Within the Sm domain, Mja-Hfq bears a conserved “Hfq Sm2 motif” (FKHAI) and the loop residues that form the A/U-binding pockets in S. aureus Hfq are also well conserved in the archaeal protein (Schumacher et al. 2002). Collectively, these findings suggest that an Hfq-like protein may be capable of functionally replacing an archaeal Lsm protein. It is therefore conceivable that insight into the function of the M. jannaschii protein could further the understanding of Lsm function in Archaea. To this end, we have undertaken a structural and functional analysis of the Mja-Hfq protein. We here report its crystal structure at 2.5 Å resolution and provide evidence that archaeal and bacterial Hfqs have a number of biochemical and biological properties in common. Specifically, we show that Mja-Hfq binds efficiently to E. coli sRNAs and that the protein possesses the ability to promote intermolecular base-pairing. In addition, Mja-Hfq promotes translation of the general stress sigma factor σS in E. coli and can complement, at least partially, a number of phenotypes displayed by an E. coli hfq knockout strain.
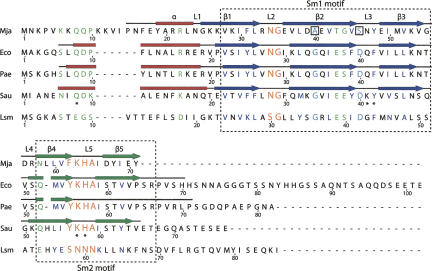
FIGURE 1.
Hfq sequence alignment. Alignment of the sequence of Mja-Hfq (Mja) with those bacteria homologous for which a structure has been determined, E. coli (Eco), P. aeruginosa (Pae), and S. aureus (Sau), as well as the Saccharomyces cerevisiae Sm-like (Lsm) protein. The location of the Sm1 and Sm2 sequence motifs is indicated by dashed boxes, and the secondary structure observed in the four structures is indicated above its corresponding sequence, with helices (α) as red boxes and sheets (β1–β5) as dark blue (Sm1) or green (Sm2) arrows. The loops (L1–L5) are shown as lines. In the sequence alignment, partly conserved residues or residues with conserved functionality are shown in green. Highly conserved hydrophobic residues found in all Sm/Lsm proteins are indicated with dark blue letters and residues in loops L3 and L5 that form the nucleotide-binding pockets in the structure of the S. aureus Hfq–RNA complex are labeled with a star. Two of the most conserved residues amongst Hfq and Sm proteins, a glycine (G) and an aspartic acid (D) residue within the Sm1 domain (indicated with cyan), are replaced by alanine (A) and serine (S), respectively, in Mja-Hfq (boxed). The two Hfq signature motifs, the Sm1 NG and Sm2 Y(F)KHA motif, are shown with orange letters. The alignment figure was produced using SecSeq (D.E. Brodersen, unpubl. software, http://www.bioxray.au.dk/∼deb/secseq).
RESULTS
Expression, isolation, and crystallization of Mja-Hfq
The Mja-Hfq protein was overexpressed in E. coli as an N-terminal chitin–intein fusion and purified to homogeneity after on-column removal of the affinity tag. When expressed in a wild-type strain it was found that the endogenous Eco-Hfq copurified with the archaeal protein. Consequently, the Mja-Hfq protein was overexpressed in an E. coli Δhfq strain.
The purified protein readily yielded crystals belonging to the space group P21 diffracting to at least 2.5 Å resolution. The structure was determined by molecular replacement using the hexamer from the P. aeruginosa Hfq structure as a search model. The single significant solution that resulted from this calculation revealed that Mja-Hfq possesses the Sm-fold and forms a hexameric ring like its bacterial orthologs (Fig. 2A). There are two hexamers in the crystallographic asymmetric unit (ASU), and the 12 subunits are very similar with a root-mean-square deviation (RMSD) <0.7 Å for all corresponding Cα atoms (Fig. 2B). The two hexamers in the ASU are arranged side-by-side and rotated 45° relative to each other, thereby producing a fairly open packing (Supplemental Fig. 1; http://www.bioxray.au.dk/∼deb/downloads/suppmat/Nielsen_et_al_2007_Suppl_Fig_1.pdf). Pairwise comparison of the backbone fold of Mja-Hfq with the previously determined bacterial Hfq structures illustrates the close structural homology between the archaeal and the bacterial proteins (Fig. 2C).
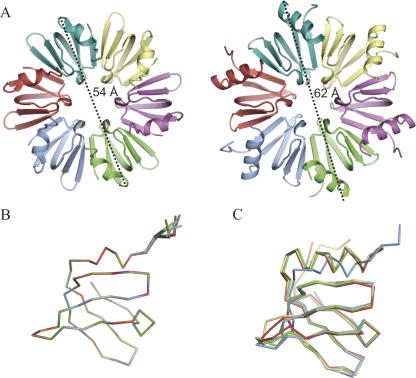
FIGURE 2.
Overview of the Mja-Hfq structure. (A) Views of the proximal side of the M. jannaschii (left) and E. coli (PDB ID 1HK9, right) Hfq hexamers. The maximum diameters of the two rings, 54 Å and 62 Å, respectively, are indicated. (B) Superposition of all 12 monomers of Mja-Hfq present in the ASU. (C) Superposition of the structures of Eco-Hfq (red), Pae-Hfq (yellow), and Sau-Hfq (green) onto Mja-Hfq (blue). The figures were prepared using PyMOL (DeLano 2002).
Significant structural differences between the archaeal and the bacterial Hfqs are restricted to the N-terminal α-helix, the variable L4 loop, and the C terminus. Specifically, the N-terminal α-helix is much shorter in Mja-Hfq, reducing it to a single-turn 3(10) helix, which in the superimposition is placed between the first and second turn of the α-helix in the bacterial Hfq proteins (Fig. 2C). This structural difference stretches the loop following the helix, which is placed much closer to the β-sheet in M. jannaschii than seen in the structures of the bacterial Hfq proteins. Consequently, the overall diameter of the hexameric Mja-Hfq ring is significantly smaller than its orthologs in bacteria (54 Å versus 62 Å) (Fig. 2A). Another structural property that sets Mja-Hfq apart from the bacterial counterparts is the charge distribution on the distal face of the ring (i.e., the face not containing the base-binding pockets), which is predominantly negative in Mja-Hfq (Fig. 3A, right) and positive in the Hfq structures of Eco-Hfq and Pae-Hfq (Fig. 3A; Sauter et al. 2003; Nikulin et al. 2005; Brennan and Link 2007). However, a positively charged distal face is not a common feature of all bacterial Hfq structures determined so far, e.g., in Sau-Hfq, where this face is predominantly nonpolar (Schumacher et al. 2002).
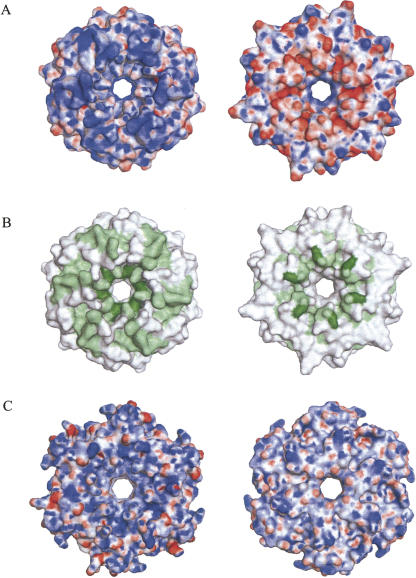
FIGURE 3.
Surface properties of Hfq. (A) Electrostatic potential mapped onto the surface of Mja-Hfq as seen from the proximal (left) and distal (right) sides. Dark blue corresponds to +25 kbT/ec and dark red to –25 kbT/ec. Surface potentials were calculated with APBS (Adaptive Poisson–Boltzmann Solver) (Baker et al. 2001) and visualized using PyMOL (DeLano 2002). (B) Conserved residues in the alignment of Mja-Hfq with the bacterial homologs mapped on the surface of Mja-Hfq in the same views as in A. Dark-green residues are identical, light-green residues are conserved in >70% of the sequences and white residues are conserved in <70% of the sequences. (C) Surface potential observed for the bacterial Hfq from E. coli (PDB ID 1HK9) in the proximal (left) and distal (right) views.
Mapping the phylogenetic conservation of Hfq residues onto the Mja-Hfq hexamer structure shows that most of the conserved residues cluster around the cationic pore region and the rim of the proximal face (Fig. 3B). Evidence that several of these conserved residues are involved in RNA binding is provided from previous work (for review, see Brennan and Link 2007). First, residues that participate in forming the A/U nucleotide-binding pockets in the Sau-Hfq-(AU5G) crystal structure spiral around the pore on the proximal surface. Second, mutational analyses of Eco-Hfq indicate that additional conserved residues on the proximal face are involved in RNA binding. Specifically, Hfq point mutations at Lys3 or Arg17 result in a protein that cannot support Qβ replication, and residue Arg16 has been implicated in binding of the regulatory DsrA RNA (Brescia et al. 2003; Sonnleitner et al. 2004; Sun and Wartell 2006). In addition, data from mutational analyses indicate the presence of another binding site on the distal face of the Eco-Hfq hexamer that specifically recognizes poly(A) RNA (Mikulecky et al. 2004; Sun and Wartell 2006). Consistently, substitution of Tyr25, Ile30, or Lys31 by alanine strongly reduced the affinity of Hfq for a 27-nucleotide (nt) synthetic poly(A) oligomer. Based on the similarities in sequence and structure, it is likely that Mja-Hfq possesses cognate sites for RNA binding on the proximal face, while the poly(A)-binding site on the distal face may not be retained in the archaeal protein. For example, the structural counterpart of the poly(A)-binding residues Ile30 and Lys31 in E. coli Hfq are Glu36 and Val37 in Mja-Hfq (Fig. 1). Furthermore, the very acidic nature of this face of the Mja-Hfq hexamer would disfavor RNA binding.
Functional analysis of Mja-Hfq
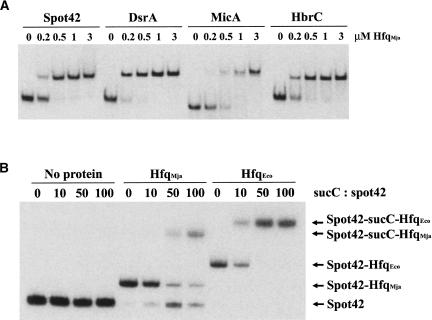
To investigate whether archaeal and bacterial Hfq also display the same biochemical and biological properties, we carried out a number of in vitro and in vivo experiments. First, we examined the RNA-binding properties of the purified Mja-Hfq protein complex by testing its binding to three well-studied small RNA regulators, Spot 42, DsrA, and MicA, as well as to a less-characterized sRNA, HbrC (Møller et al. 2002a; Brescia et al. 2003; Lease and Woodson 2004; Mikulecky et al. 2004; Rasmussen et al. 2005; Udekwu et al. 2005; Sun and Wartell 2006; P. Valentin-Hansen, unpubl.). Samples containing a fixed amount of 32P-labeled sRNA, a 500-fold excess of tRNA, and increasing amounts of Mja-Hfq protein were incubated, and complex formation was analyzed by electrophorectic mobility shift assays (Fig. 4A). The results show that the Mja-Hfq protein is capable of interacting with the sRNAs in a manner very similar to that reported for Eco-Hfq (Møller et al. 2002a; Brescia et al. 2003; Rasmussen et al. 2005); however, its affinity for the sRNAs was slightly lower (two- to sixfold) than that of the bacterial protein (data not shown). Next, we investigated whether Mja-Hfq possesses the ability to form stable ternary complexes with RNAs that function through base pairing. For this we used Spot 42 RNA, which binds to the galK and sucC translational regions of the gal- and suc-operon transcripts, respectively, in a Hfq-dependent manner (Møller et al. 2002b; C. Udesen and P. Valentin-Hansen, unpubl.). The results of the interaction studies with Spot 42 and sucC RNA are presented in Figure 4B. When a constant amount of radiolabeled Spot 42 RNA was incubated with 10-, 50-, and 100-fold molar excess of unlabeled sucC-RNA, no complex formation was observed. Addition of either Mja-Hfq or Eco-Hfq, however, efficiently triggered formation of Spot 42-Hfq-sucC complexes. For Mja-Hfq, the addition of sucC-RNA resulted in unbound Spot 42 RNA as well (Fig. 4B). The reason for this is not clear, but RNA pairing could potentially destabilize the protein–RNA interactions in the case of Mja-Hfq, which binds less efficiently to the RNAs, leading to release of the RNA. Apart from this disparity in binding behavior, the results clearly show that the archaeal protein is capable of forming ternary Hfq:mRNA:sRNA complexes.
FIGURE 4.
RNA-binding by Mja-Hfq in vitro. (A) Mja-Hfq binding to specific sRNAs from E. coli. 32P-labeled transcript of the sRNAs (4 nM final concentration) and 2 μM of tRNA were incubated with increasing amounts of the Mja-Hfq protein and complex formation was monitored in an electrophoretic mobility-shift experiment. (B) Mja-Hfq facilitates RNA–RNA interaction in vitro. Incubation of 32P-labeled Spot 42 RNA (5 nM), 2 μM tRNA, and increasing amounts of unlabeled suc′-RNA substrate in the absence (lanes 1–4) or presence of either 2 μM Mja-Hfq (lanes 5–8) or 2 μM Eco-Hfq (lanes 9–12). Complex formation was monitored in an electrophoretic mobility shift experiment. Unbound Spot 42 RNA and the various complexes formed are indicated next to the gel.
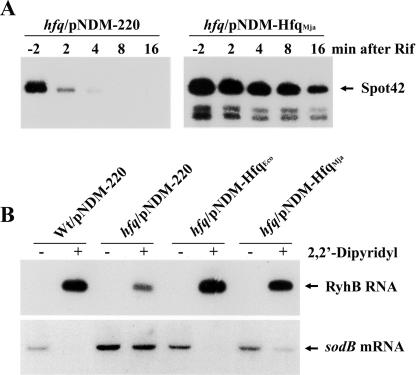
In order to study the function of the Mja-Hfq protein in vivo, we generated a low-copy-number plasmid, pNDM-hfqMja, expressing the M. jannaschii hfq gene under the control of the inducible lac promoter derivative, PA1/O4 (Lanzer and Bujard 1988). The competence of Mja-Hfq to support stabilization of sRNA was tested in rifampicin run-off experiments in IPTG-induced, exponentially grown E. coli hfq1∷kan cells carrying pNDM-hfqMja or the empty vector pNDM-220 (Gotfredsen and Gerdes 1998). The results for Spot 42 RNA are presented in Figure 5A and show that the presence of Mja-Hfq significantly increased the lifetime of the sRNA (approximately eightfold). Taken together with the in vitro results (Fig. 4A), this finding strongly indicates that the archaeal protein binds to Spot 42 RNA in vivo, thereby protecting the regulatory RNA against cleavage by endonucleases.
FIGURE 5.
Function of Mja-Hfq in E. coli in vivo. (A) Stabilization of Spot 42 RNA by Mja-Hfq. Exponential grown cultures of IPTG-induced (1 mM) cells of SØ928 hfq1∷kan carrying either pNDM-220 (control) or pNDM-hfqMja were treated with rifampicin to block new transcription. Samples were taken at the indicated times and total RNA was extracted. Spot 42 RNA levels were analyzed by Northern blot analysis. (B) Mja-Hfq cooperates in sRNA-mediated regulation of mRNA turnover. Cells of wild-type (SØ928) carrying pNDM-220 (empty vector) and the hfq1∷kan mutant transformed with pNDM-200, pNDM-hfqEco, or pNDM-hfqMja were grown in LB medium containing 1 mM IPTG at 37°C. At an OD600 of 0.4, each of the cultures was split and 2,2′-dipyridyl was added to one culture. After 10 min, samples were harvested, total RNA was extracted, and RyhB RNA and sodB mRNA levels were analyzed by Northern blot analysis.
Next, we examined whether the Mja-Hfq protein could participate in sRNA-mediated regulation of mRNA turnover in E. coli. To address this question, we investigated regulation by the RyhB RNA, which down-regulates the synthesis of a set of iron proteins by destabilizing their mRNAs. The transcription of RyhB RNA is controlled negatively by the ferric uptake repressor (Fur) protein that acts with ferrous iron as cofactor, and therefore is inactive under conditions of iron starvation. In vivo, RyhB is unstable in the absence of Hfq and the protein is required for RyhB activity (Massé and Gottesman 2002; Massé et al. 2003). In the experiment presented in Figure 5B, the effect of RyhB induction by an iron chelator (2,2′-dipyridyl) on the sodB mRNA encoding iron superoxide dismutase was analyzed by Northern blotting of total RNA isolated from a wild-type strain and from IPTG-induced hfq1∷kan cells carrying pNDM-220, pNDM-hfqEco, or pNDM-hfqMja, respectively. The data show that Mja-Hfq stabilizes the RyhB RNA, and that the protein can function in RyhB-mediated degradation of the sodB mRNA in E. coli.
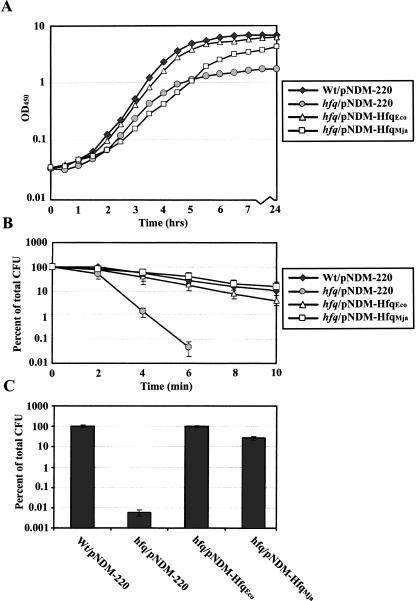
E. coli hfq knockout strains display pronounced pleiotropic phenotypes such as decreased growth rate and increased sensitivity to a number of stress factors (Tsui et al. 1994). To further examine the function of the Mja-Hfq protein, we attempted to complement the growth defect and the sensitivity of the E. coli hfq1∷kan mutant to acid and high temperature. The growth curves of the wild-type and the hfq1∷kan mutant carrying pNDM-220, pNDMhfqEco, or pNDMhfqMja, respectively, in LB medium supplemented with IPTG are presented in Figure 6A. In accordance with previous work, the growth rate, as well as the optical densities at stationary phase, were reduced for the hfq mutant strain compared with the wild type (Tsui et al. 1994). As would be expected, these growth defects could be restored upon ectopic expression of E. coli Hfq. Also, expression of Mja-Hfq restored the stationary growth defect (final yield) of the mutant strain; however, the growth rate of the mutant was not affected by the presence of the archaeal protein. The phenotype studies further revealed that ectopic expression of Mja-Hfq caused clear effects on thermotolerance and resistance to acid in the hfq1 mutant (Fig. 6B,C). In fact, the resistance of the hfq1 strain carrying pNDMhfq Mja to these types of stress was almost comparable to that found for the wild type.
FIGURE 6.
Complementation of hfq phenotypes by Mja-Hfq. (A) Growth phenotype. Growth curves of the parent strain (SØ928) carrying pNDM-220 (empty vector) and the hfq1::kan mutant transformed with pNDM-220, pNDM-hfqEco, or pNDM-hfqMja in LB medium containing 1 mM IPTG and 30 μg/mL ampicillin. (B) Thermotolerance of stationary-phase cells; 0.1 mL samples of overnight cultures of the four strains were incubated at 55°C for 0–10 min and then placed on ice. The number of colony-forming units (CFU)/mL of cultures was determined by plating dilutions on LB plates containing 30 μg/mL ampicillin and 1 mM IPTG and counting colonies overnight incubation. (C) Acid stress resistance of stationary-phase cells. Samples of overnight cultures of the four strains were diluted 100-fold in either LB medium (control) or acidic LB (pH 3, adjusted with HCl) and then incubated 1 h at 37°C. Surviving colony-forming units were determined as described in B. The growth and stress experiments were performed three times with similar results.
The stress phenotypes studied above are highly dependent on the general stress σ factor, σS. Translation of the rpoS mRNA is known to be dramatically decreased in cells lacking Hfq, and so far, three Hfq-binding regulatory RNAs—DsrA, OxyS, and RprA—have emerged as key regulators of rpoS translation (Brown and Elliott 1996; Muffler et al. 1996; for review, see Repoila et al. 2003). These findings together with the results of Figure 6, B and C, suggest that Mja-Hfq is capable of promoting translation of the alternative σ-factor. Therefore, we directly determined the σS levels in the strains used for the phenotype studies by quantitative Western blotting. The results at different stages of growth are presented in Figure 7 and show that ectopic expression of Mja-Hfq and Eco-Hfq resulted in a similar strong increase in the σS concentration during the transition from vegetative cells to stationary cells. Moreover, the σS levels in early (T) and late stationary phase (S) of the hfq-induced strains were similar to those observed in the wild-type strain. The former strains, however, had significant higher levels of the σ factor during the log phase (EE and LE), which may be attributed to a higher expression of Hfq from the plasmid than from the chromosome (about 1.3- and twofold for Eco-Hfq in log phase and stationary phase cells, respectively). Based on these findings, we conclude that Mja-Hfq possesses the ability to promote rpoS translation in a manner similar to its bacterial counterpart.
FIGURE 7.
The Mja-Hfq protein can functionally replace E. coli Hfq in rpoS translation. The wild-type/pNDM-220 strain and the hfq1∷kan strain transformed with pNDM-220, pNDM-hfqEco, and pNDM-hfqMja, respectively, were cultivated in LB medium containing 1 mM IPTG and 30 μg/mL ampicillin. Cells were harvested at various time points after inoculation, and quantitative Western analysis was done on equal amounts of cells to determine the relative σS levels at each point during growth. (EE) Early-log cells; (LE) late-log cells; (T) cells at transition to stationary phase; (S) stationary phase cells.
DISCUSSION
Prokaryotic genomes encode a single or a maximum of three Sm-related proteins. Structural and biochemical studies of several of these proteins have provided general insight into the oligomerization behavior and RNA-binding properties of Sm-like proteins. At present, the best-studied system is Eco-Hfq and the best-characterized function of this protein is its role in mediating post-transcriptional effects of small regulatory RNAs. In contrast, we have very little information regarding the in vivo functions of the Sm-related proteins in Archaea. Here, we report the crystal structure and functional characterization of an Hfq ortholog from M. jannaschii, which is the first, and thus far, only archaeal species found to contain an hfq gene.
Our structure of the M. jannaschii protein reveals that each of its polypeptide chains adopts a fold essentially identical to that observed for its bacterial counterpart, a curved five-stranded β-sheet capped by an N-terminal α-helix and separated by a very short “variable region” (loop 4). In this regard, it is of interest that two of the most conserved residues amongst Hfq and Sm proteins, a glycine (G34) and an aspartic acid (D40) residue within the Sm1 domain, are replaced by alanine (A40) and serine (S46), respectively, in Mj-Hfq (Fig. 1, boxed in section). Gly34 is a structural component of a conserved β-bulge on the external edge of the ring, and Asp40 is a conserved hydrogen-bonded residue that forms part of the binding-site L3-loop on the inside surface of the ring. In the bacterial Hfqs, the side chain of Leu46 from β-strand 3 is approaching Gly34 (closest distance ∼3.8 Å) and the side chain of Asp40 is hydrogen bonded to the main-chain amide of Val43 (the first residue of β-strand 3). This interaction seems to help keep the binding determinants on loop 3 properly aligned in the A/U-specific pocket. In Mja-Hfq, the presence of a less-bulky residue (Val52) at the position that corresponds to Leu46 allows Ala40 to occupy the same position as Gly34 in the bacterial proteins. Furthermore, loop 3 is stabilized by a hydrogen bond between the side chains of Ser46 (S) and Glu49 (E) (Fig. 1), and this interaction is almost identical in position with that of Asp40 and Val43 in the bacterial Hfqs. Thus, the substitution of alanine for glycine and serine for aspartic acid within the Sm1 domain of the archaeal protein correlates structurally with substitutions of residues at position 49 and 52 of β-strand 3.
The overall structure of the Mja-Hfq hexamer is also very similar to its orthologs in bacteria. This reinforces the suggestion that formation of hexameric rings is a hallmark of the Hfq family of proteins. However, the N-terminal α-helix of the archaeal protein is significantly shorter than seen in bacteria, containing just a single 3(10)-type turn in contrast to the three regular α-helical turns seen in the bacterial proteins. This affects the location of the α-helix, which in the Mja-Hfq structure is situated solely on the proximal side of the hexamer, while being both on the top and on the side of the complex in the bacterial structures. Consequently, the overall diameter of the archaeal ring is not defined by the extent of the α-helix, but by the loops of the β-sheet and, therefore, is significantly smaller. The crystallographic data further demonstrate that the contacts between monomers are essentially identical to those observed for Eco-Hfq. In fact, the monomers of Eco-Hfq and Mja-Hfq can associate with each to form hetero-hexamers when Mja-Hfq is expressed in wild-type E. coli. In addition, the loop residues that form the A/U-binding pockets in the S. aureus Hfq-AU5G complex structure are conserved in the Mja-Hfq protein, indicating that an equivalent binding site is likely to be present in M. jannaschii. This assumption is fully consistent with our in vitro and in vivo experiments, demonstrating that Mja-Hfq and Eco-Hfq have similar properties with respect to sRNA and mRNA binding.
Mutational analysis indicates that there may be an additional binding region recognizing poly(A) RNA on the distal face of the Eco-Hfq hexamer (Mikulecky et al. 2004; Sun and Wartell 2006). Such a locus is unlikely to be present in the archaeal protein, since the poly(A)-binding determinants are not conserved, leading to a change in charge distribution on the distal faces of Mja-Hfq, which is predominantly negative (Fig. 3). In this regard, we note that distal face mutations of Eco-Hfq did not significantly affect DsrA RNA and rpoS mRNA 5′UTR binding. Moreover, no functional role of this binding region has been reported.
To obtain information about the function of Mja-Hfq in vivo, we investigated whether the archaeal protein can functionally replace Eco-Hfq. Most notably, our data established that Mja-Hfq possesses the ability to promote ribo-regulation by the regulatory RyhB RNA. Intriguingly, this finding suggests that the archaeal protein, in addition to facilitating RNA–RNA interaction in vivo, is capable of interacting directly with components of the mRNA decay machinery, even though the factors and pathways involved have diverged in the two lineages. Previous work established that RyhB-mediated degradation of the E. coli sodB mRNA depends on Hfq as well as on RNAse E, the major endonuclease involved in mRNA decay (Massé et al. 2003). Specifically, it was established that the destabilization of the sodB message is dependent on the C-terminal domain of RNase E, known to be responsible for the assembly of a general RNA decay machinery, called the RNA degradosome. Moreover, RyhB-mediated mRNA degradation was found to be coupled to turnover of the regulatory RNA. These results led to the suggestion that formation of RyhB–mRNA complex could promote RNA degradation by recruiting RNase E or the degradosome to the message (for review, see Carpousis 2003; Massé et al. 2003). Indeed, recent studies by Aiba and coworkers have provided experimental evidence for such a model by showing that RNase E interacts directly with Hfq, as well as with Hfq bound to small RNAs, via its C-terminal scaffold region, and these complexes are clearly distinct from those formed by the degradosome components (Morita et al. 2005). Taken together, the studies imply that formation of a specialized nucleoprotein complex, wherein Hfq acts as an adaptor between RNase E and RyhB, is required for rapid turnover of the sodB mRNA (Aiba 2007). Moreover, the ability of Mja-Hfq to promote RyhB regulation indicates that it possesses the properties of a chaperone very similar to that of Eco-Hfq. In this context, E. coli Hfq has been shown to promote intermolecular annealing of RyhB and the sodB mRNA by altering the folding of the mRNA (Geissmann and Touati 2004). Our in vivo data further establish that Mja-Hfq can complement stress phenotypes observed in E. coli hfq knockout strains that are dependent on the general stress σ factor, σS. In agreement with this, we found that Mja-Hfq is capable of promoting translation of the rpoS mRNA. However, our studies of cell growth reveal that the archaeal protein does not fully complement the growth defects of the E. coli hfq1 strain.
In summary, we have shown here that the Hfq protein of the archaeon M. jannaschii is structurally and functionally closely related to bacterial Hfq. Specific areas in which parallels can be drawn between the archaeal protein and the E. coli Hfq include binding specificity, chaperone activity, protein–protein interaction, and the ability to promote RNA–RNA interactions. Furthermore, the archaeal Hfq is capable of modulating translation and facilitating small RNA function in E. coli. It therefore seems probable that Mja-Hfq plays a role in vivo that is comparable to that of E. coli Hfq. In addition, the absence of an Lsm gene in M. jannaschii may be indicative of related functions for archaeal Hfq and Lsm proteins.
Currently, the exact evolutionary relationship between Hfq and Sm-like proteins is not clear, but it is believed that archaea and eukaryotes evolved from a common predecessor that diverted from the bacterial domain (Woese et al. 1990). In keeping with this view, the Sm domains of archaea and eukaryotes are homologous at both the sequence and structural levels, and the proteins possess similar RNA-binding properties and oligomerization behavior. In contrast, Hfq has a divergent sequence compared with the eukaryotic and archaeal Sm/Lsm proteins. Moreover, Hfq possesses a relaxed RNA-binding specificity that enables it to participate in many stages of RNA metabolism. Therefore, Hfq may represent an ancient, less-specialized form of the Sm family of proteins.
MATERIALS AND METHODS
Bacterial strains
All strains used were E. coli-K12 derivatives: SØ928 (Δdeo, Δlac) (Valentin-Hansen et al. 1978), SØ928hfq1∷kan (Rasmussen et al. 2005), ER2566 (New England Biolabs), and ER2566Δhfq (obtained from M. Reijns).
Plasmid construction
For overexpression of Mja-Hfq in E. coli, a codon-optimized gene flanked by SapI and PstI sites (5′-GCTCTTCCAACATGAACAAGCCTGTGAAGAAACAGCAACCAAAAAAGGTCATCCCGAATTTTGAATACGCGCGCCGTCTGAATGGTAAAAAAGTTAAAATTTTTCTTCGTAACGGAGAGGTGCTGGATGCCGAGGTCACCGGGGTTAGCAACTATGAAATCATGGTGAAAGTTGGCGATCGGAATTTGTTAGTTTTCAAACATGCAATAGACTATATTGAATACTAACTGCAG-3′) was synthesized by Entelechon GmbH and cloned into pTYP11 from New England Biolabs (generating pTYB11-hfqMja). Plasmid pNDM-hfqEco was constructed by replacing the EcoRI–BamHI fragment of pNDM220 with a PCR-generated fragment, prepared using EcoHfq-pNDM-F (5′-CGGGATCCAAAGGAGGAATTAACTATGGCTAAGGGGCAATCTTTACAAGATCCGTTCC-3′) and EcoHfq-pNDM-Rev (5′-CGGAATTCTTATTCGGTTTCTTCGCTGTCCTGTTGCGCGGAAGTATTCTG-3′) as primers and SØ928 chromosomal DNA as template, followed by digestion with EcoRI and BamHI. Plasmid pNDM-HfqMja was constructed by replacing the EcoRI–BamHI fragment of pNDM220 (Gotfredsen and Gerdes 1998) with a PCR-generated fragment, prepared using P4-pNDM-Bam (5′-CGGGATCCAAAGGAGGAATTAACTATGAACAAGCCTGTGAAGAAACAGCAACC-3′) and P4-pNDM-rev (5′-CGGAATTCTTAGTATTCAATATAGTCTATTGCATGTTT) as primers and plasmid pTYB11-hfqMja as template, followed by digestion with EcoRI and BamHI.
Protein purification
The E. coli Hfq protein was purified as described (Møller et al. 2002a). The full-length M. jannaschii Hfq protein was overexpressed in an E. coli Δhfq derivative of the ER2566 strain and purified using the intein system (Impact-CN TM, New England Biolabs). Cells were grown at 25°C in 2x YT medium containing 100 μg/mL ampicillin. At an OD600 of ∼0.6, induction was triggered by addition of 0.4 mM IPTG for 16 h. Cells were harvested by centrifugation and lysed using a French press. Following centrifugation, protein purification was carried out according to the manufacturer's recommendations for the intein system (New England Biolabs). The lysis and wash buffers contained 20 mM Na-HEPES (pH 8.0), 1 mM EDTA (pH 8.0), and 0.5 M or 0.8 M NaCl, respectively. On-column cleavage of the intein-fusion tag was carried out for 2 d at room temperature using a lysis buffer containing 50 mM DTT. The eluted Mja-Hfq protein was precipitated by adding ammonium sulphate to 60% saturation.
Crystallization, data collection, and processing
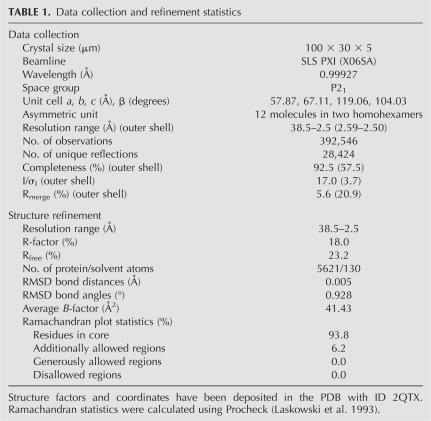
Prior to crystallization, the protein sample was run on a Superdex 200 100/30 GL gel-filtration column (GE Healthcare) to remove any aggregates and exchange the protein into crystallization buffer (20 mM Na-HEPES at pH 8.0, 200 mM NaCl, 5 mM β-mercaptoethanol). Monodisperse fractions were pooled and concentrated in a VivaSpin 500 μL spin filter (30.000 MWCO, Viva Sciences) to a final concentration of 8 mg/mL as measured by OD280 absorption (ε0 = 2 mg/mL*OD280). Crystallization screens were set up using the sitting-drop vapor-diffusion method by mixing protein solution with reservoir in 2:1 volume ratios. Initial crystallization conditions were identified using the Index Screen (Hampton Research) and subsequently optimized to 0.1 M Tris (pH 8.5), 25% PEG 3350, and 0.2 M ammonium acetate. Crystals appeared as thin plates after ∼1 wk and grew to a full size of around 100 × 30 × 5 in about 1–2 wk. Single crystals were removed from the cryo-protecting mother liquor using 100 or 200 μm litho loops (Hampton Research) and frozen directly in liquid nitrogen. Native diffraction data extending to 2.5 Å resolution were collected at PXI at the Swiss Light Source (SLS). The data were indexed and processed in HKL2000 (Otwinowski and Minor 1997) and initial phases were obtained by molecular replacement (MR) using the P. aeruginosa Hfq (pdb:1U1S) as a search model in Phaser (McCoy et al. 2005). MR was followed by solvent flattening and 12-fold NCS averaging in Resolve (Terwilliger 2003), which removed model bias and dramatically improved the electron-density map. The initial model was manually inspected and corrected according to the electron density and refined, initially with a 12-fold NCS-restraint in Refmac (Murshudov et al. 1997), subsequently in Phenix (Adams et al. 2002) using standard restraints, which improved the X-ray residuals to a final Rwork/Rfree of 18.0/23.2%. X-ray data and refinement statistics are given in Table 1. The final structure has been deposited in the PDB (www.rcsb.org) with PDB ID 2QTX.
TABLE 1.
Data collection and refinement statistics
Gel-shift analysis and in vitro synthesis of RNA
In vitro synthesis of Spot 42 and MicA (SraD) RNAs, and 32P end-labeling were carried out as described by Møller et al. (2002a,b) and Rasmussen et al. (2005). DNA templates carrying a T7 promoter were generated by PCR for synthesis of DsrA, HbrE, and the sucC′ mRNA substrate. The primers used, were DsrA RNA: T7-dsrA (5′-ATCCTGTAATACGACTCACTATTAGGACACATCAGATTTCCTGGTGTTTCA-3′) and dsrA-rev (5′-AAATCCCGACCCTGAGGGGGTCG-3′); HbrC RNA: T7-hbrC (5′-TGTAATACGACTCACTATAGATTCGTTATCAGTGCAGG-3′) and 3′-hbrC (5′-AAAAACCGCCAGTAAACC-3′); suc′RNA: T7/sucC (5′-GGGGATCCTAATACGACTCACTATAGGGAGACCGGAAGCGATACGAAATATTCGG-3′) and sucC-rev (5′-GGGAATTCGCGCGGAGTAGTACAGGC-3′).
Half-life determination and Northern blot analysis
For half-life determination of Spot 42 RNA, bacterial cultures were grown in Luria-Bertani (LB) medium at 30°C to an OD600 of 0.6 and treated with rifampicin (300 μg/mL). At the indicated times, 30-mL samples were collected by centrifugation at 0°C. Total RNA purification and Northern blot analysis were performed as described (Møller et al. 2002a,b). The 5′ end-labeled DNA probes (EM1 and EM33) used for detection of RyhB RNA and sodB mRNA are described in Massé and Gottesman (2002).
Western blot analysis of σS
Overnight cultures were diluted in LB medium containing 30 μg/mL ampicillin and 1 mM IPTG to an OD600 of 0.02 and grown at 37°C. At various time points during growth, 1-mL cell-samples were removed and equal amounts of total protein from each sample were electrophoretically separated on 12% SDS–polyacrylamide gels, and blotted onto an Immobilon-P membrane (Millipore). The membrane was probed with a 1:10.000 dilution of anti-RpoS monoclonal antibodies (NeoClone) and subsequently probed with a 1:2.000 dilution of anti-mouse alkaline phosphatase conjugate (Dako Cytomation). Western blots were developed using Western lightning chemiluminescence Reagent PLUS (PerkinElmer Life Sciences).
Acid resistance test and thermotolerance of stationary-phase cells
Stationary-phase resistance was determined for the following strains: SØ928/pNDM220, SØ928 hfq1∷kan/pNDM220, SØ928 hfq1∷kan/pNDM-HfqEco, and SØ928 hfq1∷kan/pNDM-HfqMja. Cultures were started from single colonies and grown overnight at 37°C in LB medium supplemented with 30 μg/mL ampicillin and 1 mM IPTG. For the acid tolerance experiment, the overnight cultures were diluted 1:100 in “neutral” LB (control) and acidic LB (adjusted to pH 3 with HCl), respectively, and incubated for 1 h at 37°C. The number of CFU/mL was determined by plating serial dilutions of cells onto LB agar plates containing 30 μg/mL ampicillin and 1 mM IPTG. The plates were incubated overnight at 37°C and acid survival was calculated as the percentage of cells remaining after acid treatment. For the thermotolerance experiment, 100 μL of the overnight culture was incubated at 55°C and samples removed after 0, 2, 4, 6, 8, and 10 min of heat treatment and placed on ice. Subsequently, surviving colony-forming units were determined as described for the acid-resistance test.
ACKNOWLEDGMENTS
We are grateful to Martin Reijns for providing the strain ER2566Δhfq. This work was funded by grants from the Danish Natural Science Research Council, the Danish National Research Foundation (Grundforskningsfonden), the Novo Nordisk Foundation, and a Career Development Award from the Human Frontier Science Program to D.E.B.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.689007.
REFERENCES
- Adams, P.D., Grosse-Kunstleve, R.W., Hung, L.-W., Ioerger, T.R., McCoy, A.J., Moriarty, N.W., Read, R.J., Sacchettini, J.C., Sauter, N.K., Terwilliger, T.C. PHENIX: Building new software for automated crystallographic structure determination. Acta Crystallogr. 2002;D58:1948–1954. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- Aiba, H. Mechanism of RNA silencing by Hfq-binding small RNAs. Curr. Opin. Microbiol. 2007;10:134–139. doi: 10.1016/j.mib.2007.03.010. [DOI] [PubMed] [Google Scholar]
- Baker, N.A., Sept, D., Joseph, S., Holst, M.J., McCammon, J.A. Electrostatics of nanosystems: Application to microtubules and the ribosome. Proc. Natl. Acad. Sci. 2001;98:10037–10041. doi: 10.1073/pnas.181342398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beggs, J. Lsm proteins and RNA processing. Biochem. Soc. Trans. 2005;33:433–438. doi: 10.1042/BST0330433. [DOI] [PubMed] [Google Scholar]
- Brennan, R.G., Link, T.M. Hfq structure, function and ligand binding. Curr. Opin. Microbiol. 2007;10:125–133. doi: 10.1016/j.mib.2007.03.015. [DOI] [PubMed] [Google Scholar]
- Brescia, C.C., Mikulecky, P.J., Feig, A.L., Sledjeski, D.D. Identification of the Hfq-binding site on DsrA RNA: Hfq binds without altering DsrA secondary structure. RNA. 2003;9:33–43. doi: 10.1261/rna.2570803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown, L., Elliott, T. Efficient translation of the RpoS σ factor in Salmonella typhimurium requires host factor I, an RNA-binding protein encoded by the hfq gene. J. Bacteriol. 1996;178:3763–3770. doi: 10.1128/jb.178.13.3763-3770.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpousis, A.J. Degradation of targeted mRNAs in Escherichia coli: Regulation by a small antisense RNA. Genes & Dev. 2003;17:2351–2355. doi: 10.1101/gad.1147003. [DOI] [PubMed] [Google Scholar]
- Collins, B.M., Harrop, S.J., Kornfeld, G.D., Dawes, I.W., Curmi, P.M.G., Mabbutt, B.C. Crystal structure of a heptameric Sm-like protein complex from archaea: Implications for the structure and evolution of snRNPs. J. Mol. Biol. 2001;309:915–923. doi: 10.1006/jmbi.2001.4693. [DOI] [PubMed] [Google Scholar]
- Cooper, M., Johnston, L.H., Beggs, J.D. Identification and characterization of Uss1p (Sdb23p): A novel U6 snRNA-associated protein with significant similarity to core proteins of small nuclear ribonucleoproteins. EMBO J. 1995;14:2066–2075. doi: 10.1002/j.1460-2075.1995.tb07198.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Haseth, P.L., Uhlenbeck, O.C. Interaction of Escherichia coli host factor protein with oligoriboadenylates. Biochemistry. 1980;19:6138–6146. doi: 10.1021/bi00567a029. [DOI] [PubMed] [Google Scholar]
- DeLano, W.L. The PyMOL molecular graphics system. 2002 http://www.pymol.org
- Geissmann, T.A., Touati, D. Hfq, a new chaperoning role: Binding to messenger RNA determines access for small RNA regulator. EMBO J. 2004;23:396–405. doi: 10.1038/sj.emboj.7600058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gotfredsen, M., Gerdes, K. The Escherichia coli relBE genes belong to a new toxin–antitoxin gene family. Mol. Microbiol. 1998;29:1065–1076. doi: 10.1046/j.1365-2958.1998.00993.x. [DOI] [PubMed] [Google Scholar]
- Gottesman, S. The small RNA regulators of Escherichia coli: Roles and mechanisms. Annu. Rev. Microbiol. 2004;58:303–328. doi: 10.1146/annurev.micro.58.030603.123841. [DOI] [PubMed] [Google Scholar]
- Hermann, H., Fabrizio, P., Raker, V.A., Foulaki, K., Hornig, H., Brahms, K., Lührmann, H. snRNP Sm proteins share two evolutionarily conserved sequence motifs which are involved in Sm protein–protein interactions. EMBO J. 1995;14:2076–2088. doi: 10.1002/j.1460-2075.1995.tb07199.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kambach, C., Walke, S., Young, R., Avis, J.M., de la Fortelle, E., Raker, V.A., Lührmann, R., Li, J., Nagai, K. Crystal structures of two Sm protein complexes and their implications for the assembly of the spliceosomal snRNPs. Cell. 1999;96:375–387. doi: 10.1016/s0092-8674(00)80550-4. [DOI] [PubMed] [Google Scholar]
- Kawamoto, H., Koide, Y., Morita, T., Aiba, H. Base-pairing requirement for RNA silencing by a bacterial small RNA and acceleration of duplex formation by Hfq. Mol. Microbiol. 2006;61:1013–1022. doi: 10.1111/j.1365-2958.2006.05288.x. [DOI] [PubMed] [Google Scholar]
- Khusial, P., Plaag, R., Zieve, G.W. LSm proteins form heptameric rings that bind to RNA via repeating motifs. Trends Biochem. Sci. 2005;30:522–528. doi: 10.1016/j.tibs.2005.07.006. [DOI] [PubMed] [Google Scholar]
- Kilic, T., Thore, S., Suck, D. Crystal structure of an archaeal Sm protein from Sulfolobus solfataricus . Proteins. 2005;61:689–693. doi: 10.1002/prot.20637. [DOI] [PubMed] [Google Scholar]
- Lanzer, M., Bujard, H. Promoters largely determine the efficiency of repressor action. Proc. Natl. Acad. Sci. 1988;85:8973–8977. doi: 10.1073/pnas.85.23.8973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laskowski, R.A., MacArthur, M.W., Moss, D.S., Thornton, J.M. PROCHECK: A program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 1993;26:283–291. [Google Scholar]
- Lease, R.A., Woodson, S.A. Cycling of the Sm-like protein Hfq on the DsrA small regulatory RNA. J. Mol. Biol. 2004;344:1211–1223. doi: 10.1016/j.jmb.2004.10.006. [DOI] [PubMed] [Google Scholar]
- Massé, E., Gottesman, S. A small RNA regulates the expression of genes involved in iron metabolism in Escherichia coli . Proc. Natl. Acad. Sci. 2002;99:4620–4625. doi: 10.1073/pnas.032066599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massé, E., Escorcia, F.E., Gottesman, S. Coupled degradation of a small regulatory RNA and its mRNA targets in Escherichia coli . Genes & Dev. 2003;17:2374–2383. doi: 10.1101/gad.1127103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCoy, A.J., Grosse-Kunstleve, R.W., Storoni, L.C., Read, R.J. Likelihood-enhanced fast translation functions. Acta Crystallogr. 2005;D61:458–464. doi: 10.1107/S0907444905001617. [DOI] [PubMed] [Google Scholar]
- Mikulecky, P.J., Kaw, M.K., Brescia, C.C., Takach, J.C., Sledjeski, D.D., Feig, A.L. Escherichia coli Hfq has distinct interaction surfaces for DsrA, rpoS, and poly(A) RNAs. Nat. Struct. Mol. Biol. 2004;11:1206–1214. doi: 10.1038/nsmb858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Møller, T., Franch, T., Højrup, P., Keene, D.R., Bächinger, H.P., Brennan, R.G., Valentin-Hansen, P. Hfq: A bacterial Sm-like protein that mediates RNA-RNA interaction. Mol. Cell. 2002a;9:23–30. doi: 10.1016/s1097-2765(01)00436-1. [DOI] [PubMed] [Google Scholar]
- Møller, T., Franch, T., Udesen, C., Gerdes, K., Valentin-Hansen, P. Spot 42 RNA mediates discoordinate expression of the E. coli galactose operon. Genes & Dev. 2002b;16:1696–1706. doi: 10.1101/gad.231702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita, T., Maki, K., Aiba, H. RNase E-based ribonucleoprotein complexes: Mechanical basis of mRNA destabilization mediated by bacterial noncoding RNAs. Genes & Dev. 2005;19:2176–2186. doi: 10.1101/gad.1330405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muffler, A., Fischer, D., Hengge Aronis, R. The RNA-binding protein HF-I, known as a host factor for phage Q β RNA replication, is essential for rpoS translation in Escherichia coli . Genes & Dev. 1996;10:1143–1151. doi: 10.1101/gad.10.9.1143. [DOI] [PubMed] [Google Scholar]
- Muffler, A., Traulsen, D.D., Fischer, D., Lange, R., Hengge Aronis, R. The RNA-binding protein HF-I plays a global regulatory role which is largely, but not exclusively, due to its role in expression of the σs subunit of RNA polymerase in Escherichia coli . J. Bacteriol. 1997;179:297–300. doi: 10.1128/jb.179.1.297-300.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mura, C., Cascio, D., Sawaya, M.R., Eisenberg, D.S. The crystal structure of a heptameric archaeal Sm protein: Implications for the eukaryotic snRNP core. Proc. Natl. Acad. Sci. 2001;98:5532–5537. doi: 10.1073/pnas.091102298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mura, C., Kozhukhovsky, A., Gingery, M., Phillips, M., Eisenberg, D. The oligomerization and ligand-binding properties of Sm-like archaeal proteins (SmAPs) Protein Sci. 2003;12:832–847. doi: 10.1110/ps.0224703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murshudov, G.N., Vagin, A.A., Dodson, E.J. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. 1997;D53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- Nikulin, A., Stolboushkina, E., Perederina, A., Vassilieva, I., Blaesi, U., Moll, I., Kachalova, G., Yokoyama, S., Vassylyev, D., Garber, M., et al. Structure of Pseudomonas aeruginosa Hfq protein. Acta Crystallogr. D Biol. Crystallogr. 2005;61:141–146. doi: 10.1107/S0907444904030008. [DOI] [PubMed] [Google Scholar]
- Nogueira, T., Springer, M. Post-transcriptional control by global regulators of gene expression in bacteria. Curr. Opin. Microbiol. 2000;3:154–158. doi: 10.1016/s1369-5274(00)00068-0. [DOI] [PubMed] [Google Scholar]
- Otwinowski, Z., Minor, W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- Pannone, B.K., Wolin, S.L. RNA degradation: Sm-like proteins wRING the neck of mRNA. Curr. Biol. 2000;10:R478–R481. doi: 10.1016/S0960-9822(00)00552-2. [DOI] [PubMed] [Google Scholar]
- Rasmussen, A.A., Eriksen, M., Gilany, K., Udesen, C., Franch, T., Petersen, C., Valentin-Hansen, P. Regulation of ompA mRNA stability: The role of a small regulatory RNA in growth phase-dependent control. Mol. Microbiol. 2005;58:1421–1429. doi: 10.1111/j.1365-2958.2005.04911.x. [DOI] [PubMed] [Google Scholar]
- Repoila, F., Majdalani, N., Gottesman, S. Small noncoding RNAs, coordinators of adaptation processes in Escherichia coli: The RpoS paradigm. Mol. Microbiol. 2003;48:855–861. doi: 10.1046/j.1365-2958.2003.03454.x. [DOI] [PubMed] [Google Scholar]
- Sauter, C., Basquin, J., Suck, D. Sm-like proteins in Eubacteria: The crystal structure of the Hfq protein from Escherichia coli . Nucleic Acids Res. 2003;31:4091–4098. doi: 10.1093/nar/gkg480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schumacher, M.A., Pearson, R.F., Møller, T., Valentin-Hansen, P., Brennan, R.G. Structures of the pleiotropic translational regulator Hfq and an Hfq-RNA complex: A bacterial Sm-like protein. EMBO J. 2002;21:3546–3556. doi: 10.1093/emboj/cdf322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senear, A.W., Steitz, J.A. Site-specific interaction of Qß host factor and ribosomal protein S1 with Qß and R17 bacteriophage RNAs. J. Biol. Chem. 1976;251:1902–1912. [PubMed] [Google Scholar]
- Séraphin, B. Sm and Sm-like proteins belong to a large family: Identification of proteins of the U6 as well as the U1, U2, U4, and U5 snRNPs. EMBO J. 1995;14:2089–2098. doi: 10.1002/j.1460-2075.1995.tb07200.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnleitner, E., Moll, I., Bläsi, U. Functional replacement of the Escherichia coli hfq gene by the homologue of Pseudomonas aeruginosa . Microbiol. 2002;148:883–891. doi: 10.1099/00221287-148-3-883. [DOI] [PubMed] [Google Scholar]
- Sonnleitner, E., Napetschnig, J., Afonyushkin, T., Ecker, K., Vecerek, B., Moll, I., Kaberdin, V.R., Bläsi, U. Functional effects of variants of the RNA chaperone Hfq. Biochem. Biophys. Res. Commun. 2004;323:1017–1023. doi: 10.1016/j.bbrc.2004.08.190. [DOI] [PubMed] [Google Scholar]
- Storz, G., Opdyke, J.A., Zhang, A.X. Controlling mRNA stability and translation with small, noncoding RNAs. Curr. Opin. Microbiol. 2004;7:140–144. doi: 10.1016/j.mib.2004.02.015. [DOI] [PubMed] [Google Scholar]
- Sun, X., Wartell, R.M. Escherichia coli Hfq binds A18 and DsrA domain II with similar 2:1 Hfq6/RNA stoichiometry using different surface sites. Biochemistry. 2006;45:4875–4887. doi: 10.1021/bi0523613. [DOI] [PubMed] [Google Scholar]
- Sun, X.G., Zhulin, I., Wartell, R.M. Predicted structure and phyletic distribution of the RNA-binding protein Hfq. Nucleic Acids Res. 2002;30:3662–3671. doi: 10.1093/nar/gkf508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terwilliger, T.C. SOLVE and RESOLVE: Automated structure solution and density modification. Methods Enzymol. 2003;374:22–36. doi: 10.1016/S0076-6879(03)74002-6. [DOI] [PubMed] [Google Scholar]
- Thore, S., Mayer, C., Sauter, C., Weeks, S., Suck, D. Crystal structures of the Pyrococcus abyssi Sm core and its complex with RNA. J. Biol. Chem. 2003;278:1239–1247. doi: 10.1074/jbc.M207685200. [DOI] [PubMed] [Google Scholar]
- Törö, I., Thore, S., Mayer, C., Basquin, J., Séraphin, B., Suck, D. RNA binding in an Sm core domain: X-ray structure and functional analysis of an archaeal Sm protein complex. EMBO J. 2001;20:2293–2303. doi: 10.1093/emboj/20.9.2293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Törö, I., Basquin, J., Teo-Dreher, H., Suck, D. Archaeal Sm proteins form heptameric and hexameric complexes: Crystal structures of the Sm1 and Sm2 proteins from the hyperthermophile Archaeoglobus fulgidus . J. Mol. Biol. 2002;320:129–142. doi: 10.1016/S0022-2836(02)00406-0. [DOI] [PubMed] [Google Scholar]
- Tsui, H.C.T., Leung, H.C.E., Winkler, M.E. Characterization of broadly pleiotropic phenotypes caused by an Hfq insertion mutation in Escherichia coli-K-12. Mol. Microbiol. 1994;13:35–49. doi: 10.1111/j.1365-2958.1994.tb00400.x. [DOI] [PubMed] [Google Scholar]
- Udekwu, K.I., Darfeuille, F., Vogel, J., Reimegård, J., Holmqvist, E., Wagner, E.G. Hfq-dependent regulation of OmpA synthesis is mediated by an antisense RNA. Genes & Dev. 2005;19:2355–2366. doi: 10.1101/gad.354405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valentin-Hansen, P., Svenningsen, B.A., Munch-Pedersen, A., Hammer-Jespersen, K. Regulation of the deo operon in Escherichia coli: The double negative control of the deo operon by the cytR and deoR repressors in a DNA directed in vitro system. Mol. Gen. Genet. 1978;159:191–202. doi: 10.1007/BF00270893. [DOI] [PubMed] [Google Scholar]
- Valentin-Hansen, P., Eriksen, M., Udesen, C. The bacterial Sm-like protein Hfq: A key player in RNA transactions. Mol. Microbiol. 2004;51:1525–1533. doi: 10.1111/j.1365-2958.2003.03935.x. [DOI] [PubMed] [Google Scholar]
- Wilusz, C.J., Wilusz, J. Eukaryotic Lsm proteins: Lessons from bacteria. Nat. Struct. Mol. Biol. 2005;12:1031–1036. doi: 10.1038/nsmb1037. [DOI] [PubMed] [Google Scholar]
- Woese, C.R., Kandler, O., Wheelis, M.L. Towards a natural system of organisms: Proposal for the domains Archaea, Bacteria, and Eucarya. Proc. Natl. Acad. Sci. 1990;87:4576–4579. doi: 10.1073/pnas.87.12.4576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, A.X., Wassarman, K.M., Ortega, J., Steven, A.C., Storz, G. The Sm-like Hfq protein increases OxyS RNA interaction with target mRNAs. Mol. Cell. 2002;9:1–22. doi: 10.1016/s1097-2765(01)00437-3. [DOI] [PubMed] [Google Scholar]