Abstract
Within the past years RNA interference (RNAi) has become one of the most valuable tools for post-transcriptional gene silencing. Making RNAi temporally and/or spatially controllable would even enlarge its scope of application. Attaching a light-removable protection group to siRNAs is a very promising approach to achieve this control over RNAi. It has been reported that modifying siRNA nucleobases surrounding the mRNA cleavage site between the 10th and 11th nucleotides successfully suppresses RNAi. We investigated the influence of photolabile protection groups at these and the adjacent nucleobases on siRNA activity and chose to incorporate caged deoxynucleotides instead of ribonucleotides. The siRNAs designed by these means were shown to be completely inactive. By irradiation with UV light (366 nm) they could be fully reactivated and showed the same activity as their unmodified siRNA counterparts.
Keywords: RNAi, caging, spatiotemporal control
INTRODUCTION
Since its discovery in 1998 (Fire et al. 1998), RNA interference (RNAi) has rapidly become a powerful tool for post-transcriptional gene silencing (for reviews, see, for example, McManus and Sharp 2002; Dorsett and Tuschl 2004; Meister and Tuschl 2004). RNAi is believed to be an ancient mechanism of cells to defend themselves against invasion of viral RNA. It leads to a degradation of those mRNAs that are partly complementary to the antisense strand of short, dsRNA oligonucleotides—the so-called small interfering RNAs (siRNA) (Hamilton and Baulcombe 1999). This siRNA can originate from an insert encoding for transcription of long double-stranded or short hairpin RNAs (dsRNA, shRNA), which are then processed to siRNAs by the endonuclease Dicer (Bernstein et al. 2001). Alternatively, mature siRNAs can be directly introduced into the cell. There the siRNA binds to a protein complex called the RNA-induced silencing complex (RISC), for which a free 5′-phosphate in the siRNA is required (Chiu and Rana 2002). The stability of the 5′ base-pairing is crucial for the decision concerning which strand of the siRNA remains within the RISC and serves as a template for mRNA recognition (Schwarz et al. 2003). mRNA degradation is a catalytic process. One single siRNA can induce the cleavage of several copies of mRNA (Kennerdell and Carthew 1998), which are always cut at the position opposite of the 10th and 11th nucleotides from the siRNA's 5′ end (Elbashir et al. 2001).
Normally the RNAi machinery is initiated as soon as siRNAs enter the cell, and the first down-regulatory effects can be observed soon after transfection. However, many interesting applications become possible if the spatial pattern of RNAi can be controlled or if a precisely defined starting point of knock-down can be chosen that differs from the moment of transfection. Hence some efforts have been made to make RNAi spatiotemporally inducible.
One approach to realizing a temporal control is to use shRNA transcripts and set their expression under the control of an externally addressable promoter, as in a drug-controllable promoter-based system (for review, see Wiznerowicz et al. 2006). Still, these systems have some problems that need to be solved, like an insufficient knock-down in the induced state in some cases as summarized by Wiznerowicz (see, for example, Berns et al. 2004).
A second approach is to transfect an inactive siRNA, which can be subsequently activated inside the cell. This can be achieved by transfecting temporarily inactivated “caged” siRNAs that have been modified with a photolabile protection group (for a review on caging, see, for example, Mayer and Heckel 2006). In a first attempt Friedman et al. did a random caging of the siRNA phosphate backbone (Shah et al. 2005). In this study, functioning siRNAs could be successfully inactivated and later reactivated after irradiation with UV light. The problem with this approach was that either the inactivation or the activation was not complete. Fully inactive siRNA could not be entirely activated and vice versa.
A more precise strategy using just one single caging group is blocking the 5′-phosphate of siRNA (Nguyen et al. 2006; Shah and Friedman 2007). Inhibiting this group prevents siRNAs from binding to the RISC and therefore prohibits RNAi (Chiu and Rana 2002; Czauderna et al. 2003). Even though in this approach fully active siRNAs were obtained after irradiation, a residual activity of caged siRNAs remained.
Another vulnerable spot of siRNA with key functionality is the central region surrounding the point of mRNA scission. It has been shown previously that modifying a single nucleotide in this central part of an siRNA, leading to a bulge in the double helix, completely disrupts RNAi (Chiu and Rana 2003).
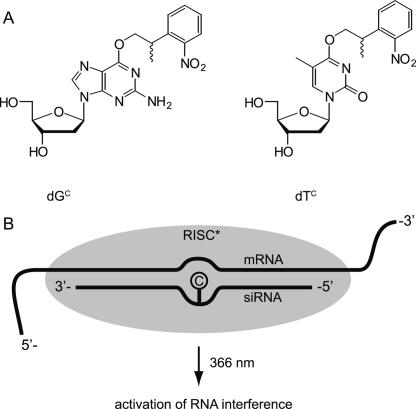
We introduced a photolabile modification in this region in an siRNA targeting enhanced green fluorescent protein (EGFP). Instead of ribonucleotides we inserted modified deoxyribonucleotides, as they are well tolerated at these positions (Chiu and Rana 2003). This choice represents a synthetic shortcut and allowed us to circumvent extra steps required for the separation of the O2/O3 regioisomers during phosphoramidite synthesis. The caging group used is the established 2-(2-nitrophenyl)propyl-group (NPP group), which we introduced at the O6 position of guanosine and the O4 position of thymine (Fig. 1A). The caging groups interfere with the hydrogen bonding pattern of the nucleobases and introduce a steric hindrance to the base pair formation. Hence these derivatives can be seen as temporary mismatches creating a bulge in the siRNA:mRNA duplex (Fig. 1B) until the photolysis event in which the caging groups are completely removed and unmodified nucleobases are formed again. In previous studies we have already shown that nucleobase-caged nucleotides can be used to trigger transcription with light, to turn protein activity either on or off, or to trigger conformational changes by using caged aptamers (Heckel and Mayer 2005; Kröck and Heckel 2005; Mayer et al. 2005; Heckel et al. 2006). Other groups used a similar approach with caged nucleobases to study RNA folding kinetics (Höbartner and Silverman 2005; Wenter et al. 2005). In our present study, as a model system we chose a dual fluorescence reporter assay with EGFP and red fluorescent protein (RFP), which we investigated in HeLa cells. What we observed is that the caged siRNA was fully inactive until cells were irradiated with UV light, which led to a complete restoration of siRNA activity.
FIGURE 1.
(A) Structure of the caged deoxynucleotides dGC and dTC used in this study. (B) The photolabile NPP group masks the Watson–Crick interaction capability of the nucleobases to create a temporary bulge region in the siRNA:mRNA duplex of the RISC and can be completely removed by irradiation with light in the near UV range.
RESULTS
Introduction of single deoxynucleotides does not affect siRNA activity
From a previous study we knew that siRNA activity can be compatible with a 2′ modification including also only hydrogen (Chiu and Rana 2003). To be sure that this also applies in our case we began our study with a series of siRNAs with single deoxynucleotides in different positions (Fig. 2). A reliable system to investigate down-regulation of genes in cell culture is the dual fluorescence reporter assay (Chiu and Rana 2002, 2003). We chose EGFP as our target and RFP for normalization, as siRNAs targeting one and not the other are commercially available (Qiagen). Both genes are under the control of the constitutively expressed CMV promoter. Together with the siRNA the plasmids were cotransfected into HeLa cells followed by analysis 24 h later. The fluorescence of EGFP was normalized to that of RFP and the fluorescence intensity of samples transfected with only plasmids and no RNA was set to 100%. The anti-EGFP siRNA obtained from Qiagen was transfected as a positive control and considered as maximum accessible down-regulation. Each experiment was performed in triplicate.
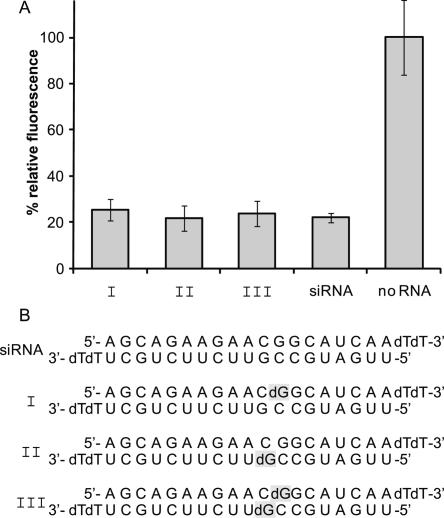
FIGURE 2.
Introduction of single deoxynucleotides does not affect siRNA activity. (A) EGFP expression of HeLa cells transfected with siRNAs that contain single deoxynucleotides within the sense strand (I), the antisense strand (II), and both strands (III) determined by measuring fluorescence intensity. EGFP fluorescence intensity was normalized to that of RFP. Fluorescence intensity of a sample that was not treated with siRNA (no RNA) was set to 100%. The down-regulation of EGFP expression in a positive control sample transfected with a commercially available siRNA was assumed to be the maximum available one. From the figure it can be seen that down-regulation of the deoxynucleotide-containing siRNA sequences (I–III) is as effective as the one of the positive control (siRNA). (B) Sequences of the siRNA strands used. The positions of the deoxynucleotides are marked in gray.
siRNAs were modified once within the sense strand (I, caged at the 12th nucleotide) and once within the antisense strand (II, caged at the 9th nucleotide), and a third sample (III) was a hybrid of the described modified sense and antisense strands bearing two modifications. In each case a guanosine was replaced with a deoxyguanosine. All siRNA sequences, modified and unmodified, are depicted in Figure 2.
As expected, the substitution of a ribonucleotide by a deoxyribonucleotide did not affect the siRNA efficiency. Down-regulation of EGFP expression with all of the tested modified sequences was within error limits identical to the down-regulation of the control siRNA.
Stability of the caging group in cell culture
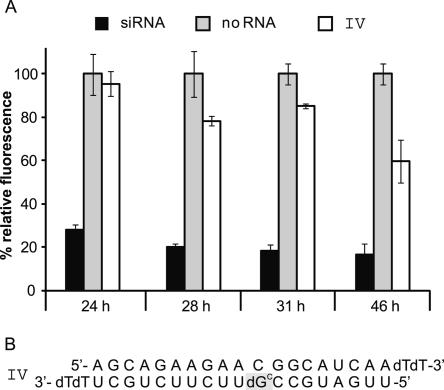
RNAi is an effect of temporary nature. While siRNA-mediated down-regulatory effects in plants and worms, for example, can, due to an amplification system, outlast several generations (Dalmay et al. 2000; Grishok et al. 2000), in mammalian cells gene silencing is only of limited duration. For HeLa cells transfected with EGFP-encoding plasmids, the RNAi effect has been reported to peak at ∼42 h past transfection. A decrease sets in about 66 h past transfection (Chiu and Rana 2002). Using the (un)caging approach the beginning of RNAi can be delayed. However, as cells have various mechanisms to correct mutations in oligonucleotides, it is conceivable that one of those mechanisms might also target the artificially introduced cage. To find out how long an initiation of siRNA activity can be deferred until the cage is eliminated, we performed another experiment. As the siRNA II with ribonucleotide–deoxyribonucleotide exchange at position 9 in the antisense strand was already successfully tested, for this preliminary experiment with a caged siRNA we chose to introduce the caged deoxynucleotide at exactly the same position (siRNA IV, Fig. 3).
FIGURE 3.
Stability of the caging group in cell culture. (A) EGFP expression after different periods of proliferation. HeLa cells were transfected with caged siRNA IV and proliferated without irradiation for increasing time spans before fluorescence intensity was analyzed. While 24 h past transfection the caged siRNA is still inactive, after 28 h a reactivation with resulting decreasing fluorescence intensity of EGFP sets in, which further progresses with increasing time to reach an activation of ∼40% after 46 h past transfection. (B) In the siRNA IV the photolabile protection group is positioned at nucleotide # 9 of the antisense strand.
HeLa cells were cultured in common 24-well plates and transfected with the mentioned reporter gene plasmids as well as a positive control (commercially obtained siRNA) and a negative control (no RNA) and samples with the caged siRNA IV (Fig. 3).
Cells proliferated for different time frames (without irradiation); lysis followed after 24, 28, 31, and 46 h past transfection. While expression of EGFP after a proliferation period of 24 h still remains at the same level as the negative control, which means that siRNA activity is still fully blocked, after 28 h a first decrease in fluorescence intensity becomes apparent. Even though this effect does not seem to be linear, as the siRNA of the 31-h proliferation sample is not more active than the sample with 3 h less cell growth, after 42 h the increase in siRNA activity further progressed and a down-regulation of 40% was measurable (Fig. 3).
Nucleobase caging at cleavage site leads to reversible inactivation of siRNA
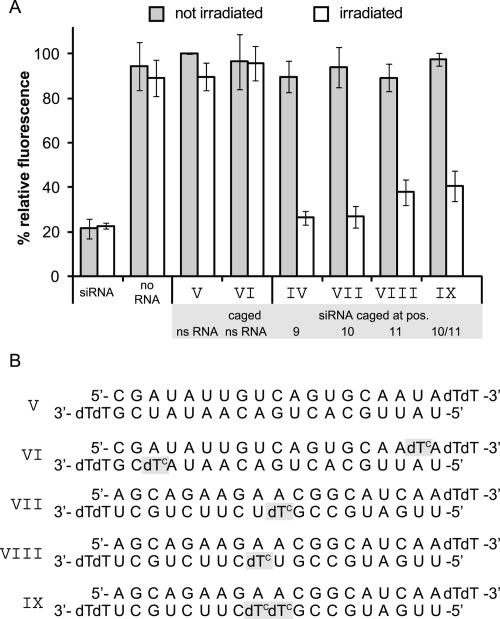
As it is likely that a perturbation at the cleavage site within the central region of the siRNA:mRNA duplex will interfere with mRNA cleavage, caging a position adjacent to nucleotides surrounding the point of mRNA scission appeared to be very promising. We had already shown that a cage in position 9 of the antisense strand (siRNA IV) prevents siRNA activity. Therefore we generated siRNA sequences where nucleotides 10 and 11 were replaced by a caged deoxynucleotide (Fig. 4B, siRNAs VII and VIII, respectively) as well as a doubly caged sequence bearing the cage at both positions 10 and 11 (Fig. 4B, siRNA IX) to check if these caged siRNAs perform even better. Additionally, as another negative control, we included a nonsilencing RNA (Fig. 4B, ns RNA V), which targets neither the expression of EGFP nor the one of RFP, and a caged variant thereof (Fig. 4B, ns RNA VI). In contrast to the previous experiments, this time the sample transfected with this nonsilencing ns RNA V was taken as standard. Its fluorescence intensity was set to 100% since the cells of this sample obtained the same amount of oligonucleotides during transfection, which should be even more precise than exclusively normalizing EGFP fluorescence of a mock-treated sample (“no RNA”) to RFP fluorescence intensity. In most of our previously quoted studies with caged nucleic acids we had used UV-LEDs, which have become available recently (Bernadelli et al. 2005). These UV-LEDs have about 100 times the power of mercury lamps and hence deprotection is very fast. For our studies in cell culture, however, we could not use these UV-LEDs due to the limitation in irradiation area. Therefore we had to use mercury lamps in these studies, which afford a longer deprotection time—knowing that on a single-cell level using a UV-LED or UV laser coupled to a microscope the deprotection will again be much faster. Irradiation followed 4 h past transfection for 40 min. An identical set of samples was prepared but not irradiated. To get a maximum fluorescence signal and down-regulation, cell proliferation time was chosen as the maximum time span within which cage cleavage does not yet set in, i.e., 24 h. The experiments were performed in triplicate and repeated several times, and the results are shown in Figure 4. The gray bars of Figure 4 show the results of the nonirradiated samples. Within error limits all newly tested sequences were as inactive as the nonsilencing 21mer RNA V, and fluorescence intensity does not deviate from that one of mock-treated cells. In one experiment a slight residual activity showed up. In this case the caged siRNA sequence had been stored for a prolonged time. The problem could be eliminated by RP-HPLC repurification, which revealed a small amount (∼3%) of already uncaged sequence. This is in accordance with observations other groups have made with impurities in their siRNA samples (Nguyen et al. 2006). Irradiation of the caged nonsilencing RNA sample VI clearly shows that no down-regulation of EGFP expression is initiated due to byproducts of uncaging. The caged siRNA sequences that have been irradiated (white bars) display a recovery of activity to the same amount as the positive control in case of siRNAs IV and VII with a cage at positions 9 and 10, while it is not fully restored in the case of caging at position 11 (siRNA VIII) and the doubly caged sequence (siRNA IX). A possible explanation for the incomplete recovery of siRNA activity of the doubly caged siRNA might be a less clean photodeprotection behavior (see, for example, Supplemental Figs. 1 and 2).
FIGURE 4.
Nucleobase caging at cleavage site leads to reversible inactivation of siRNA. (A) Fluorescence intensity of HeLa cells transfected with different siRNAs with (gray bars) and without (white bars) irradiation for 40 min 4 h past transfection. As a positive control, indicating full activity, a commercially available siRNA was transfected. The various negative controls were a sample not treated with any siRNA (no RNA), a sample transfected with a nonsilencing RNA (V) whose fluorescence intensity was defined as 100%, and the same nonsilencing RNA bearing photolabile protection groups (VI). The different caged siRNA sequences (IV, VII, VIII, and IX) are all inactive within error limits. For sequences IV and VII, a reactivation that can be assumed to be complete could be achieved, while sequences VIII and IX, even after irradiation, display a residual inactivation. Fluorescence intensity of the control samples was not affected by irradiation and achieves values similar to those of the not irradiated control samples. Also, products of uncaging do not affect EGFP expression in any way, as the fluorescence intensities of VI irradiated and not irradiated are equal. (B) Sequences and caging positions of the used 21mers. The sequences V and VI are not targeting the expression of EGFP or the normalization protein RFP.
Caging of the cleavage site-surrounding nucleobases apparently inhibits the cleavage step of RNAi
It sounds plausible that manipulating the nucleobases flanking the cleavage site of an siRNA:mRNA duplex should interfere with the mRNA cleavage step, but it is not impossible that, in fact, a different, earlier step in RNAi could be disturbed. For example, it is thinkable that a caging group might prevent siRNA from entering the RISC or disturb mRNA recognition. The latter is not too likely as it has already been shown by different groups that RNAi—and that implies mRNA recognition—tolerates even multiple mismatches, though observations about in which region the mismatches are tolerated best differ (Amarzguioui et al. 2003; Chiu and Rana 2003; Haley and Zamore 2004). To examine whether the hypothesis of inhibition of the cleavage step applies, we tested some sequences bearing cages at positions other than the central cleavage region in the siRNA, which should then still be active.
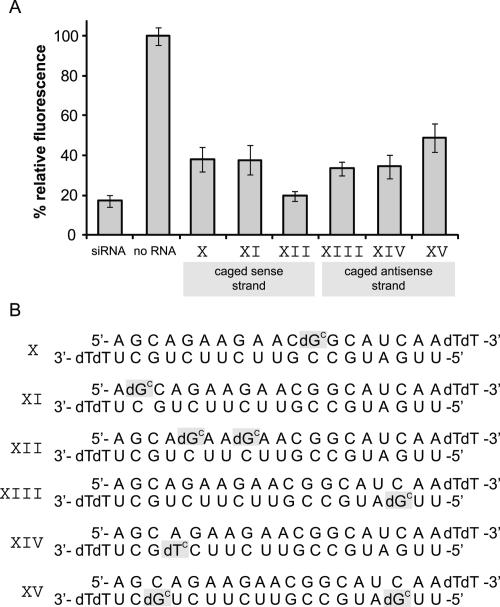
RISC initially binds double-stranded siRNA, and not until later does it release one strand. If an siRNA modification should inhibit RISC formation, such a modification within the sense strand should inhibit RNAi just as well as a caged antisense strand. Thus, we not only introduced a cage into the antisense strand (siRNAs XIII, XIV, and XV in Fig. 5), but also into the sense strand (siRNAs X, XI, and XII in Fig. 5). All of these sequences are active, though not to the same extent as the positive control. The maximum inhibition that can still be seen is ∼50% for the doubly caged antisense sequence (siRNA XV). Apparently, introduction of a cage at arbitrary bases mostly somehow interferes with the RNAi mechanism, but not nearly as effectively as if it is positioned within the cleavage region. Therefore, we still assume that the effectively caged siRNAs IV, VII, and VII work via a local duplex perturbation around the scission site in the RISC complex and thus prevent mRNA cleavage. Supplemental Figure 2 shows CD spectra of the unmodified siRNA and of siRNAs IV and VII. As expected all curves look very similar and show that the overall duplex structure remains mostly unaffected. Melting point studies reveal the expected small decrease in duplex stability (68.0 ± 0.5°C for the unmodified siRNA versus 64.8 ± 0.5°C for siRNA VII). We had shown before that such a very localized duplex perturbation can be used to trigger T7 RNA polymerase activity with light (Kröck and Heckel 2005).
FIGURE 5.
Caging of the cleavage site-surrounding nucleobases apparently inhibits the cleavage step of RNAi. (A) Fluorescence intensity of samples caged at positions other than those surrounding the cleavage site. HeLa cells were transfected with numerous siRNAs bearing one or two caging groups at positions either within the sense strand or not within the central cleavage region of the antisense strand. None of these sequences displays an inactivation close to that of the earlier tested sequences with cages within the cleavage region.
DISCUSSION
Introducing a photolabile protection group that suppresses the activity of siRNAs enhances the potential of RNAi applications in various ways. Not only does it provide the opportunity to determine the exact time for RNAi to set in, but it also allows for spatial control of the effect, as it is possible to exclusively irradiate and thereby activate the desired part of a sample. Diverse chemical moieties and positions within the siRNA sequence allow for targeting siRNA activity in different ways. Ideally, the caged siRNA should be fully inactive and uncaging should release the full silencing capability—in other words a clean on/off behavior is very desirable.
So far two different approaches of inactivating siRNAs by introduction of photolabile protection groups exist. The very first one was the protection of the phosphate backbone with di-methoxy-nitrophenyl-ethyl (DMNPE) groups; however no distinct point of the RNAi mechanism was targeted because phosphates were modified statistically (Shah et al. 2005). Even though significant inactivation and reactivation by irradiation could be achieved, only sequences with a residual activity of ∼50% could be fully activated, while activity of inactivated sequences could only be restored to an extent of ∼50%. A newer strategy exploits the fact that for integration of siRNA into RISC, an intact 5′-phosphate is essential. At this position a nitrophenyl-ethyl (NPE) group was introduced with good results: siRNA activity could be reduced to ∼40% and be fully restored by irradiation with UV light (Nguyen et al. 2006). The authors assume that a slight contamination with sequences being one nucleotide shorter and thus bearing no cage, which appear as side products during oligonucleotide synthesis, are responsible for this residual activity. They show a dependency of the amount of impurities and the residual activity of transfected siRNA. Lately another group has further investigated 5′-phosphate caging of siRNAs (Shah and Friedman 2007). Herein the conclusion is that impurities of shorter noncaged abortion sequences are not alone responsible for residual activity of 5′-caged siRNA, but rather a minor tolerance for 5′-blocked phosphates exists even though literature partly argues to the contrary (Chiu and Rana 2002; Czauderna et al. 2003).
Phosphates are not the only moieties where a caging group can be introduced. While it has already been reported that blocking of the 2′-OH group of siRNA nucleotides does not affect siRNA activity but on the contrary gives protection against nuclease degradation, single nucleobases are very sensitive to modifications. Among the individual nucleotides still there are some which display a great tolerance against mutations, but it has also been reported that others are highly mutation sensitive. So Chiu and Rana observed complete suppression of RNAi when attaching substituents to the bases of nucleotides surrounding the cleavage site, i.e., the 10th and 11th nucleotides, of an siRNA (Chiu and Rana 2003). These findings convinced us when we were thinking of introducing nucleobase-caged deoxynucleotides into the cleavage relevant region as a potential target site for perturbing RNAi. The choice to introduce caged deoxynucleotides instead of ribonucleotides was made because this avoids the effort of separating the 2′-/3′-regioisomers during RNA phosphoramidite synthesis. We found that substitution of single ribonucleotides against deoxynucleotides within this area of siRNA does not affect RNAi in any way, which is in accordance with literature (Chiu and Rana 2003). The caged siRNA sequences designed by this means showed very satisfying results. All of them were inactivated by at least 90%, averaged over multiple experiments. In single experiments each individual of the sequences reached an inactivation of >97% (data not shown). The best inactivation with an average value of ∼97% was achieved with a doubly caged siRNA; however, this sequence could not regain full activity through irradiation. Concerning reactivation, caging one of the two nucleotides upstream next to the cleavage site was most successful. It can be said that these two sequences worked best considering inactivation as well as activation capability. In accord with another group's observation (Nguyen et al. 2006), in some cases we found that impurities of already deprotected sequences (identified by RP-HPLC) in a range of ∼3% can cause eminent loss of inactivation. A more important aspect is the time up to which the caged sequences remain inactive inside cells; 28 h after transfection, a reactivation of the siRNA sets in even without irradiation. At the moment two explanations seem to be plausible. The cage might be cleaved, be it an active effect, mediated by the cell's oligonucleotide repairing system, or just a tribute to the chemical environment within the cell. It is also conceivable that, similar to the findings of Friedman for the 5′-phosphate cage, a slight tolerance toward the caging group occurs. In conclusion, we have shown that modification of an siRNA antisense strand with a caged deoxynucleotide in the region of mRNA scission affords inactive siRNAs that can be fully reactivated upon irradiation with light. This technique will be helpful, for example, in developmental biology for studies in model organisms for which only a few components involved in pattern formation are available. Recent studies with caged mRNA (Ando et al. 2001, 2005) or caged antisense oligodeoxynucleotide analogs (Shestopalov et al. 2007; Tang et al. 2007) in zebrafish underline the general usefulness of this approach.
MATERIALS AND METHODS
Preparation and characterization of oligonucleotides
Positive control siRNAs were obtained from Qiagen. All other siRNA oligonucleotide single strands were chemically synthesized on an ABI 392-8 DNA synthesizer using standard phosphoramidites from AZCO. Phosphoramidites carrying an NPP group were synthesized as described before (Kröck and Heckel 2005; Mayer et al. 2005). For the deprotection, oligonucleotides were treated with an 1:1 mixture of 33% MeNH2 in H2O and 41% MeNH2 in EtOH or 4 h (unmodified or caged dG) or in a 3:1 mixture of 29% NH3 (aq.) in EtOH for 15 h (caged dT) at room temperature, separated from the solid supports, evaporated, and treated with 1 M TBAF in THF at room temperature. Upon addition of Tris·HCl buffer (pH 7.4) the reaction was stopped followed by desalting on NAP columns (Amersham Biosciences). Purification was performed by anion exchange HPLC (AE-HPLC) followed by RP-HPLC to remove already uncaged oligonucleotides. AE-HPLC was performed on a DNAPac PA 100 column (4 × 250 mm, dionex), flow 1.5 mL/min, elution at 80°C; eluent A 25 mM Tris·HCl (pH 8.0), eluent B 25 mM Tris•HCl (pH 8.0), 0.5 M NaClO4. RP-HPLC was performed on a Nucleosil 100-5 C 18 (4.6 × 250 mm; CS Chromatographie Service), 1 mL/min, elution at 55°C; eluent A 0.1 M NH4OAc, eluent B MeCN, 0%B for 2 min, 0–50%B in 38 min. Final LC-MS analysis (Bruker esquire HCT) showed that the NPP group was still present on the modified nucleobases. Sample HPLC traces are shown in Supplemental Figures 1 and 2. Annealing of single strands was done in phosphate buffered saline (PBS) by heating to 80°C for 1 min and subsequent cooling to room temperature for at least 30 min. Melting points were determined with a Perkin Elmer Lambda 2S and a PTP-1 peltier system and UV-Winlab 2.25.04. CD spectra (Supplemental Fig. 3) were taken using a JASCO J-810. The samples were dissolved in 1× PBS and seven scans were averaged.
Cell culture, transfection, and irradiation of cells
Adherent HeLa cells were cultivated at 37°C in Dulbecco's modified Eagle's medium (DMEM, Gibco) containing 10% fetal calf serum (FCS) and glutamine and passaged regularly. Cells were seeded to 24- or 12-well plates at subconfluence 16–20 h prior to transfection. All transfection experiments were performed as triplicates using Metafectene (Biontex) as transfecting agent according to the manufacturer's advice. Each well of a 24-well plate was provided with 200 ng of pEGFP-N1 (Clonetec), 400 ng of pDsRed Express (Clonetec), and 25 pmol of 21mer (siRNA or nonsilencing RNA) except the no-RNA control, which obtained no RNA. Twelve well plates were transfected with double the amount of all oligonucleotides. Irradiation of cells was performed with a MinUvis Hg low pressure lamp (see Supplemental Fig. 4), 4 h post-transfection for a time span of 40 min.
Cell lysis and reporter gene assay
Twenty-four hours past transfection (=20 h past irradiation) cells were washed twice with PBS, then reporter lysis buffer (Promega) was added. Afterward cells were frozen at −80°C and thawed twice, then centrifuged and the supernatant used for the fluorescence assay.
All fluorescence experiments were measured on a varioskan fluorescence spectrometer (ThermoElectron) at room temperature. EGFP fluorescence was excited at a wavelength of 488 nm and detected at 507 nm, RFP fluorescence was excited at a wavelength of 560 nm and detected at 583 nm, each with a bandwidth of 5 nm. Intensity of EGFP fluorescence was normalized to RFP fluorescence. Fluorescence intensity of a sample, which is transfected with an equal amount of nonsilencing RNA that is neither targeting pEGFP-N1 nor pDsRed Express (designed with Qiagen algorithm available at the Qiagen website) is set to 100%.
SUPPLEMENTAL DATA
All Supplemental Materials are available on the author's homepage, http://heckel.chemie.uni-bonn.de, or upon request from the author; e-mail: heckel@uni-frankfurt.de.
ACKNOWLEDGMENTS
The work has been made possible by an Emmy Noether Fellowship of the DFG for A.H. We thank Dr. M. Hafner and Dr. A. Schmitz for helpful discussions.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.753407.
REFERENCES
- Amarzguioui, M., Holen, T., Babaie, E., Prydz, H. Tolerance for mutations and chemical modifications in a siRNA. Nucleic Acids Res. 2003;31:589–595. doi: 10.1093/nar/gkg147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ando, H., Furuta, T., Tsien, R.Y., Okamoto, H. Photomediated gene activation using caged RNA/DNA in zebrafish embryos. Nat. Genet. 2001;28:317–325. doi: 10.1038/ng583. [DOI] [PubMed] [Google Scholar]
- Ando, H., Kobayashi, M., Tsubokawa, T., Uyemura, K., Furuta, T., Okamoto, H. Lhx2 mediates the activity of Six3 in zebrafish forebrain growth. Dev. Biol. 2005;287:456–468. doi: 10.1016/j.ydbio.2005.09.023. [DOI] [PubMed] [Google Scholar]
- Bernardelli, Y., Haeberli, C., Chatton, J.Y. Flash photolysis using a light emitting diode: An efficient, compact, and affordable solution. Cell Calcium. 2005;37:565–572. doi: 10.1016/j.ceca.2005.03.001. [DOI] [PubMed] [Google Scholar]
- Berns, K., Hijmans, E.M., Mullenders, J., Brummelkamp, T.R., Velds, A., Heimerikx, M., Kerkhoven, R.M., Madiredjo, M., Nijkamp, W., Weigelt, B., et al. A large-scale RNAi screen in human cells identifies new components of the p53 pathway. Nature. 2004;428:431–437. doi: 10.1038/nature02371. [DOI] [PubMed] [Google Scholar]
- Bernstein, E., Caudy, A.A., Hammond, S.M., Hannon, G.J. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature. 2001;409:363–366. doi: 10.1038/35053110. [DOI] [PubMed] [Google Scholar]
- Chiu, Y.-L., Rana, T.M. RNAi in human cells: Basic structural and functional features of small interfering RNA. Mol. Cell. 2002;10:549–561. doi: 10.1016/s1097-2765(02)00652-4. [DOI] [PubMed] [Google Scholar]
- Chiu, Y.-L., Rana, T.M. siRNA function in RNAi: A chemical modification analysis. RNA. 2003;9:1034–1048. doi: 10.1261/rna.5103703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czauderna, F., Fechtner, M., Dammes, S., Aygun, A., Klippel, A., Pronk, G.J., Giese, K., Kaufmann, J. Structural variations and stabilising modifications of synthestic siRNA in mammalian cells. Nucleic Acids Res. 2003;31:2705–2716. doi: 10.1093/nar/gkg393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalmay, T., Hamilton, A., Rudd, S., Angell, S., Baulcombe, D.C. An RNA-dependent RNA polymerase gene in Arabidopsis is required for posttranscriptional gene silencing mediated by a transgene but not by a virus. Cell. 2000;101:543–553. doi: 10.1016/s0092-8674(00)80864-8. [DOI] [PubMed] [Google Scholar]
- Dorsett, Y., Tuschl, T. siRNAs: Applications in functional genomics and potential as therapeutics. Nat. Rev. Drug Discov. 2004;3:318–329. doi: 10.1038/nrd1345. [DOI] [PubMed] [Google Scholar]
- Elbashir, S.M., Martinez, J., Patkaniowska, A., Lendeckel, W., Tuschl, T. Functional anatomy of siRNAs for mediating efficient RNAi in Drosophila melanogaster embryo lysate. EMBO J. 2001;20:6877–6888. doi: 10.1093/emboj/20.23.6877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fire, A., Xu, S., Montgomery, M.K., Kostas, S.A., Driver, S.E., Mello, C.C. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans . Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- Grishok, A., Tabara, H., Mello, C.C. Genetic requirements for inheritance of RNAi in C. elegans . Science. 2000;287:2494–2497. doi: 10.1126/science.287.5462.2494. [DOI] [PubMed] [Google Scholar]
- Haley, B., Zamore, P.D. Kinetic analysis of the RNAi enzyme complex. Nat. Struct. Mol. Biol. 2004;11:599–606. doi: 10.1038/nsmb780. [DOI] [PubMed] [Google Scholar]
- Hamilton, A.J., Baulcombe, D.C. A species of small antisense RNA in posttranscriptional gene silencing in plants. Science. 1999;286:950–952. doi: 10.1126/science.286.5441.950. [DOI] [PubMed] [Google Scholar]
- Heckel, A., Mayer, G. Light-regulation of aptamer activity: An anti-thrombin aptamer with caged thymidine nucleobases. J. Am. Chem. Soc. 2005;127:822–823. doi: 10.1021/ja043285e. [DOI] [PubMed] [Google Scholar]
- Heckel, A., Buff, M.C.R., Raddatz, M.-S.L., Müller, J., Pötzsch, B., Mayer, G. An anticoagulant with light-triggered antidote activity. Angew. Chem. Int. Ed. Engl. 2006;45:6748–6750. doi: 10.1002/anie.200602346. [DOI] [PubMed] [Google Scholar]
- Höbartner, C., Silverman, S.K. Modulation of RNA tertiary folding by incorporation of caged nucleotides. Angew. Chem. Int. Ed. Engl. 2005;44:7305–7309. doi: 10.1002/anie.200502928. [DOI] [PubMed] [Google Scholar]
- Kennerdell, J.R., Carthew, R.W. Use of dsRNA-mediated genetic interference to demonstrate that frizzled and frizzled 2 act in the wingless pathway. Cell. 1998;95:1017–1026. doi: 10.1016/s0092-8674(00)81725-0. [DOI] [PubMed] [Google Scholar]
- Kröck, L., Heckel, A. Photo-induced transcription using temporarily mismatched caged oligonucleotides. Angew. Chem. Int. Ed. Engl. 2005;44:471–473. doi: 10.1002/anie.200461779. [DOI] [PubMed] [Google Scholar]
- Mayer, G., Heckel, A. Biologically active molecules with a “light switch.”. Angew. Chem. Int. Ed. Engl. 2006;45:4900–4921. doi: 10.1002/anie.200600387. [DOI] [PubMed] [Google Scholar]
- Mayer, G., Mikat, V., Engeser, M., Heckel, A. Light induced formation of G-quadruplex DNA secondary structures. ChemBioChem. 2005;6:1966–1970. doi: 10.1002/cbic.200500198. [DOI] [PubMed] [Google Scholar]
- McManus, M.T., Sharp, P.A. Gene silencing in mammals by small interfering RNAs. Nat. Rev. Genet. 2002;3:737–747. doi: 10.1038/nrg908. [DOI] [PubMed] [Google Scholar]
- Meister, G., Tuschl, T. Mechanisms of gene silencing by double-stranded RNA. Nature. 2004;431:343–349. doi: 10.1038/nature02873. [DOI] [PubMed] [Google Scholar]
- Nguyen, Q.N., Chavli, R.V., Marques, J.T., Conrad P.G., II, Wang, D., He, W., Belisle, B.E., Zhang, A., Pastor, L.M., Witney, F.R., et al. Light controllable siRNAs regulate gene supression and phenotypes in cells. Biochim. Biophys. Acta. 2006;1758:394–404. doi: 10.1016/j.bbamem.2006.01.003. [DOI] [PubMed] [Google Scholar]
- Schwarz, D.S., Hutvágner, G., Du, T., Xu, Z., Aronin, N., Zamore, P.D. Asymmetry in the assembly of the RNAi enzyme complex. Cell. 2003;115:199–208. doi: 10.1016/s0092-8674(03)00759-1. [DOI] [PubMed] [Google Scholar]
- Shah, S., Friedman, S.H. Tolerance of RNA interference toward modifications of the 5′ antisense phosphate of small interfering RNA. Oligonucleotides. 2007;17:35–43. doi: 10.1089/oli.2006.0067. [DOI] [PubMed] [Google Scholar]
- Shah, S., Rangarajan, S., Friedman, S.H. Light activated RNA interference. Angew. Chem. Int. Ed. Engl. 2005;44:1328–1332. doi: 10.1002/anie.200461458. [DOI] [PubMed] [Google Scholar]
- Shestopalov, I.A., Sinha, S., Chen, J.K. Light-controlled gene silencing in zebrafish embryos. Nat. Chem. Biol. 2007;3:650–651. doi: 10.1038/nchembio.2007.30. [DOI] [PubMed] [Google Scholar]
- Tang, X., Maegawa, S., Weinberg, E.S., Dmochowski, I.J. Regulating gene expression in zebrafish embryos using light-activated, negatively charged peptide nucleic acids. J. Am. Chem. Soc. 2007;129:11000–11001. doi: 10.1021/ja073723s. [DOI] [PubMed] [Google Scholar]
- Wenter, P., Fürtig, B., Hainard, A., Schwalbe, H., Pitsch, S. Kinetics of photoinduced RNA refolding by real-time NMR spectroscopy. Angew. Chem. Int. Ed. Engl. 2005;44:2600–2603. doi: 10.1002/anie.200462724. [DOI] [PubMed] [Google Scholar]
- Wiznerowicz, M., Szulc, J., Trono, D. Tuning silence: Conditional systems for RNA interference. Nat. Methods. 2006;3:682–688. doi: 10.1038/nmeth914. [DOI] [PubMed] [Google Scholar]