Abstract
Cyproterone acetate (CPA) is a steroidal antiandrogen used clinically in the treatment of prostate cancer. Compared with steroidal agonists for the androgen receptor (AR) (e.g. dihydrotestosterone, R1881), CPA is bulkier in structure and therefore seemingly incompatible with the binding pockets observed in currently available x-ray crystal structures of the AR ligand-binding domain (LBD). We solved the x-ray crystal structure of the human AR LBD bound to CPA at 1.8 Å in the T877A variant, a mutation known to increase the agonist activity of CPA and therefore facilitate purification and crystal formation of the receptor·drug complex. The structure demonstrates that bulk from the 17α-acetate group of CPA induces movement of the Leu-701 side chain, which results in partial unfolding of the C-terminal end of helix 11 and displacement of the loop between helices 11 and 12 in comparison to all other AR LBD crystal structures published to date. This structural alteration leads to an expansion of the AR binding cavity to include an additional pocket bordered by Leu-701, Leu-704, Ser-778, Met-780, Phe-876, and Leu-880. Further, we found that CPA invokes transcriptional activation in the L701A AR at low nanomolar concentrations similar to the T877A mutant. Analogous mutations in the glucocorticoid receptor (GR) and progesterone receptor were constructed, and we found that CPA was also converted into a potent agonist in the M560A GR. Altogether, these data offer information for structure-based drug design, elucidate flexible regions of the AR LBD, and provide insight as to how CPA antagonizes the AR and GR.
The AR2 is a ligand-inducible steroid hormone receptor involved in regulation of prostate growth, spermatogenesis, and bone and muscle mass. Ligands that block the actions of androgens are used clinically for the treatment of prostate cancer and include the steroidal antiandrogen, CPA (Cyprostat™), and the nonsteroidal antiandrogens flutamide (Eulexin™) and bicalutamide (Casodex™). Mutations in the AR often occur in patients being treated with these compounds that increase their agonist activity and thus result in drug resistance (1-7). Paradoxically, discontinuation of drug therapy at this point may lead to tumor regression known as antiandrogen withdrawal syndrome. A number of antiandrogen withdrawal syndrome mutations occurs in regions of direct contact with the ligand that compensate for the increased bulkiness of antagonist compared with agonist compounds. Such mutations allow antagonist ligands to invoke the same AR LBD conformation as the agonist-bound AR LBD (1, 8, 9).
Molecular modeling studies have offered a number of potential docking solutions to explain how drugs more bulky than steroidal agonists can be accommodated within the AR (10-14). Our recent report of the crystal structure of the AR LBD complexed to S-3-(4-fluorophenoxy)-2-hydroxy-2-methyl-N-[4-nitro-3-(trifluoromethyl)phenyl] propionamide (hereafter referred to as S-1 as in Ref. 15), an aryl propionamide selective androgen receptor modulator that acts as an agonist for the AR, demonstrated that movement of Trp-741 provides the additional space necessary for the compound to bind (8). Similarly, CPA is seemingly too large a molecule to fit within the AR binding pocket due to its bulky substituents at the 17-position distinguishing it from steroidal AR agonists. The LNCaP prostate cancer cell line contains an AR with the T877A point mutation, which allows CPA and hydroxyflutamide (HF) to act as agonists for the AR, whereas bicalutamide remains an antagonist. Agonist activity of CPA observed in the PR may be attributed to the presence of a cysteine (Cys-891) instead of a threonine at this position. However, CPA still does not appear compatible with the binding pocket observed in the T877A AR (16) or the PR. The estrogen receptor and GR have been shown to assume different conformations when liganded to agonists and antagonists (9, 17) due to the ability of antagonists to partially unfold the LBD of the receptor. X-ray crystal structures demonstrating changes in protein folding of the AR LBD when complexed to different ligands however are not currently available. Here we report structural evidence that CPA binding to the T877A AR induces unraveling of the C terminus of helix 11 and displacement of the loop between helices 11 and 12. Based on these findings, we show that the AR mutation, L701A, confers agonist activity to CPA similar to T877A and that a similar mutation in the GR converts CPA into a potent agonist. Furthermore, identification of the expanded binding pocket occupied by CPA provides insight for exploiting receptor interactions using structure-based drug design.
MATERIALS AND METHODS
Cloning, Expression, and Purification
An AR-LBD-(663–919) was obtained by PCR amplification from a full-length AR expression construct (pCMVhAR; generously provided by Dr. Donald J. Tindall, Mayo Clinic and Mayo Foundation, Rochester, MN) with primers containing flanking restriction sites and inserted into the pGEX6P-1 plasmid vector (Amersham Biosciences). Mutations were created in the pGEX6P1-AR-(663–919), pCMVhAR, pCMVhGR, and pCMVhPR via the Stratagene QuikChange mutagenesis kit according to the manufacturer's instructions. AR LBD expression and purification were performed essentially as previously described (1). The AR LBD was expressed as a glutathione S-transferase (GST) fusion protein in Escherichia coli BL21(DE3) at 15 °C for 16 h by induction with 30 μm isopropyl 1-thio-β-d-galactopyranoside. Cells were lysed in a buffer containing 150 mm NaCl, 50 mm Tris, pH 8.0, 5 mm EDTA, 10% glycerol, 1 mg/ml lysozyme, 10 units/ml DNase I, 10 mm MgCl2, 10 mm DTT, 0.5% CHAPS, 100 μm CPA (Sigma), and 100 μm phenylmethylsulfonyl fluoride by 3 cycles of freeze thaw. The supernatant from ultracentrifugation was incubated for 1 h at 4°C with glutathione-Sepharose (Amersham Biosciences) and washed with 150 mm NaCl, 50 mm Tris, pH 8.0, 5 mm EDTA, 10% glycerol, 10 mm ATP, 100 μm CPA, 0.1% n-octyl-β-glucoside, and 1 mm DTT. The GST-LBD fusion protein was cleaved in a buffer containing 150 mm NaCl, 50 mm Tris, pH 7.0, 10% glycerol, 100 μm CPA, 0.1% n-octyl-β-glucoside, 1 mm DTT, and 5 units/mg of protein PreScission protease (Amersham Biosciences) at 4 °C overnight releasing the AR LBD from the glutathione-Sepharose resin. The supernatant was then diluted 3-fold in 10 mm Hepes, pH 7.2, 10% glycerol, 100 μm CPA, 0.1% n-octyl-β-glucoside, and 1 mm DTT and loaded onto an HP SP cation exchange-column (Amersham Biosciences). Protein was eluted with a gradient of 50 to 500 mm NaCl in the same dilution buffer. The buffer was exchanged in a Millipore 10-kDa cut-off concentrator to a buffer containing 150 mm Li2SO4, 50 mm Hepes, pH 7.2, 10% glycerol, 100 μm CPA, 0.1% n-octyl-β-glucoside, and 10 mm DTT, and protein was concentrated to 8 mg/ml.
Crystallization, Data Collection, and Structure Determination
AR LBD crystals formed in 1–2 days using the hanging drop vapor diffusion method in reservoirs containing 0.1 m Hepes, pH 7.5, and 0.5 m to 0.8 m sodium citrate in drops of equal amounts of protein and reservoir solution. Crystals grew to maximum size within 2 weeks. Prior to flash freezing in liquid nitrogen, crystals were transferred to a solution consisting of 0.1 m Hepes, pH 7.5, 0.7 m sodium citrate, and 20% ethylene glycol. Diffraction data were collected using a Rigaku RU300 rotating anode generator and an R-axis IV++ image plate (Rigaku, The Woodlands, TX), and processed with Crystal Clear software (Molecular Structure Corp., The Woodlands, TX). The x-ray crystal structure of the WT AR LBD in complex with the nonsteroidal propionamide selective androgen receptor modulator, S-1 (8) (PDB code 2AXA), was used as a starting structure for refinement using the Crystallography & NMR System (18). After an initial round of refinement, electron density maps allowed for accurate fitting of the ligand. Model building and water molecules were added using the program O (19), and further rounds of refinement were performed using rigid body, torsion angle simulated annealing, and individual temperature factor modules of the Crystallography & NMR System. The figures were prepared with PyMOL.3
Cotransfection Assay
CV-1 cells (green monkey kidney cells) were grown in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum, 2 mm glutamine, and 1% streptomycin and penicillin to confluency in 75-cm2 tissue culture flasks. Cells were then transfected with 3 μg of the CMVhAR, CMVhGR, or CMVhPR expression vectors (WT or mutant), 30 μg of a luciferase reporter construct (pMMTV-luc, generously provided by Dr. Ronald Evans at The Salk Institute, San Diego, CA), and 30 μg of a β-galactosidase expression construct (pSV-β-galactosidase, Promega, Madison, WI) via 20 μl of Lipofectamine 2000 (Invitrogen) in serum-free Dulbecco's modified Eagle's medium. After 4 h, the medium was exchanged to Dulbecco's modified Eagle's medium supplemented with 0.2% fetal bovine serum and 2 mm glutamine. Cells were transferred 12 h later to 24-well tissue culture plates and after another 6 h were treated with 0.1–1000 nm, or no drug (control). After another 24 h, cells were washed twice with cold phosphate-buffered saline and harvested by incubation with 100 μl of passive lysis buffer (Promega) for 30 min. An aliquot (50 μl) of the lysate was then added to an opaque 96-well plate, and luciferase activity was monitored after automated injection of 50 μl of luciferase substrate (Promega) with a MicroLumat-Plus LB96V luminometer (Berthold Technologies, Oak Ridge, TN) using the WinGlow software package. An aliquot (50 μl) of the lysate was also added to a clear 96-well plate along with 50 μl of β-galactosidase assay buffer. An absorbance measurement at 420 nm was taken following a 2-h incubation at 37 °C on a Dynex MRX plate reader. Luciferase activity was normalized with β-galactosidase activity to account for differences in cell number and/or loss in transfection efficiency. Transcriptional activation was interpreted as the -fold increase (relative luciferase units) over control (i.e. non-drug treated) wells.
Docking Studies
Structures were prepared for docking using Sybyl 7.1 (Tripos Inc., St. Louis, MO) by removing the ligand and water molecules, adding polar hydrogens, and assigning Kollman united atom charges. Ligands were extracted from PDB files and modified with the Sketch Molecule tool in Sybyl, and Gasteiger-Marsili charges were assigned. AutoDock3.05 (20) was used to perform docking simulations using default parameters with a grid centered around the x, y, and z coordinates of 29, 0, and 4 Å in the WT·DHT (PDB code 1I37), T877A·DHT (PDB code 1I38), and T877A·CPA complexes. The solutions with the lowest free energy of binding of 10 clusters were reported.
RESULTS
T877A-AR LBD·CPA Complex
The final model of the T877A AR LBD·CPA crystal structure at 1.8-Å resolution consists of a single monomer of AR residues 671–919, which includes 2027 protein atoms in the asymmetric unit. Electron density maps for AR residues 886–890 in the T877A·CPA were weak and significantly displaced from the starting model (S-1-bound WT AR LBD). The backbone of these amino acids was fit using the 2Fo − Fc map at the 0.6 σ level, but side chains were omitted in the model. The model also includes 134 water molecules. CPA was fit unambiguously in a single conformation using the 2Fo − Fc map contoured at the 1.0 σ level. The refined temperature factors for the atoms of CPA were similar to those of surrounding protein atoms indicating full occupancy of the ligand. Crystallography statistics are listed in Table 1.
TABLE 1.
Crystallographic data and refinement statistics
| PDB code | 2Oz7 |
| Space group | P212121 |
| Unit cell | |
| a | 55.50 |
| b | 65.82 |
| c | 71.51 |
| Resolution range (Å) | 21.96–1.80 (1.86–1.80)a |
| Number of unique reflections | 24,416 |
| Average redundancy | 2.96 (2.19) |
| Completeness % | 96.2 (71.8) |
| Rmergeb | 0.075 (0.298) |
| I/σ | 8.8 (3.1) |
| R-factorc | 0.253 |
| Rfree | 0.270 |
| r.m.s.d. bonds (Å) | 0.006 |
| r.m.s.d. angles | 1.1 |
| Mean B-value (Å2) | 28.5 |
Values for data in the last resolution shell are shown in parenthesis.
Rmerge = Σ|Ih − |〈I〉h|ΣIh, where 〈I〉h is average intensity over symmetry equivalents.
R-factor = Σ|Fobs − Fcalc|ΣFobs. The free R-factor is calculated from 10% of the reflections that are omitted from the refinement.
Binding Conformation of CPA to the T877A AR
A hydrogen bond network is present in the x-ray crystal structure between the 3-keto group of CPA with Arg-752, Gln-711, and a water molecule (see Fig. 2a). The chlorine atom on the C6-position is accommodated in a groove surrounded by hydrophobic residues of helices 4, 7, and 11. The oxygen atom of the β-acetyl group branched from the 17-position of CPA is oriented for a hydrogen bond with the nitrogen δ2 of Asn-705, whereas the methyl of this acetyl group denoted C-21 (Fig. 1) on CPA is located only 3.2 Å from Ala-877. No hydrophilic interactions are available for the 17α-acetate group of CPA, which binds in a cavity bordered by Leu-701, Leu-704, Met-780, Phe-876, and Leu-880. The backbone oxygen of Ser-778 is located <4 Å from the C-24 methyl group of the CPA acetate, but the side chain of this residue faces away from the ligand to form a hydrogen bond with the backbone nitrogen of Met-780.
FIGURE 2. Structure of the T877A AR LBD complexed to CPA.
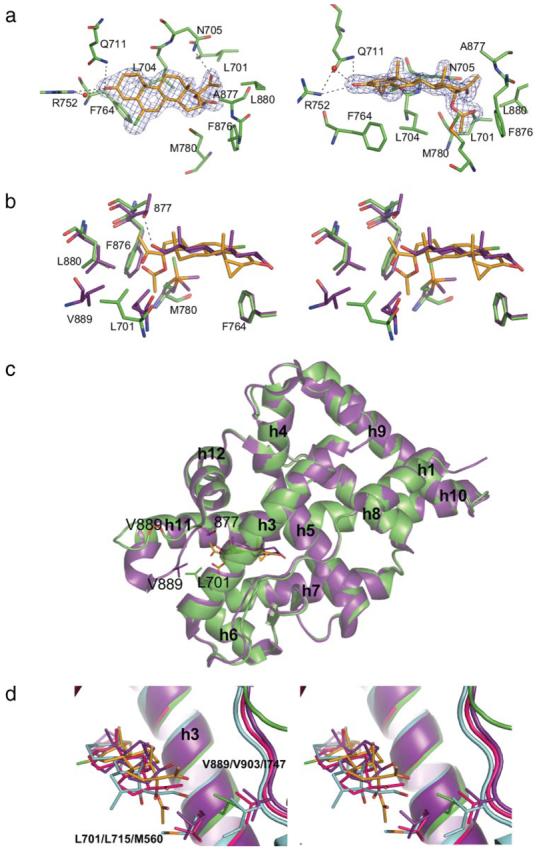
Gold, CPA carbon; green, AR carbon; red, oxygen; blue, nitrogen; green, chlorine; and orange, sulfur. a, CPA shown within the 2Fo − Fc map (blue) contoured at the 1.5 σ level at multiple angles. Note the hydrogen bonds between the 3-keto and 17β-acetyl groups of CPA. b, overlay in stereo of the WT·DHT complex (purple) (PDB code 1T7T) on the T877A·CPA structure rotated 180° about the y-axis from the right panel of a to illustrate the similar binding conformation of the steroidal molecules. Note the unfavorable proximity of the 17β-acetyl group of CPA to the Thr-877 side chain in the WT·DHT complex. Also notice the difference in the position of the Leu-701 side chain in the T877A·CPA structure from the presence of the acetate group on CPA, which is displaced into the location occupied by Val-889 in the WT·DHT complex. c, the T877A·CPA and the WT·DHT complexes superimposed illustrate the unraveling of the C terminus of helix 11 and change in the location of the loop between helices 11 and 12. d, overlay in stereo of the PR-progesterone (pink, PDB code 1A28), GR-dexamethasone (cyan, PDB code 1P93), AR·DHT (purple), and T877A AR·CPA complex. Leu-701 and Val-889 in the AR correspond to Leu-715 and Val-903 in the PR and Met-560 and Ile-747 in the GR, respectively. Notice that CPA binding to the GR and PR is likely to induce similar disruption of the helix 11 and 12 region from interaction of the helix 3 residue.
FIGURE 1.
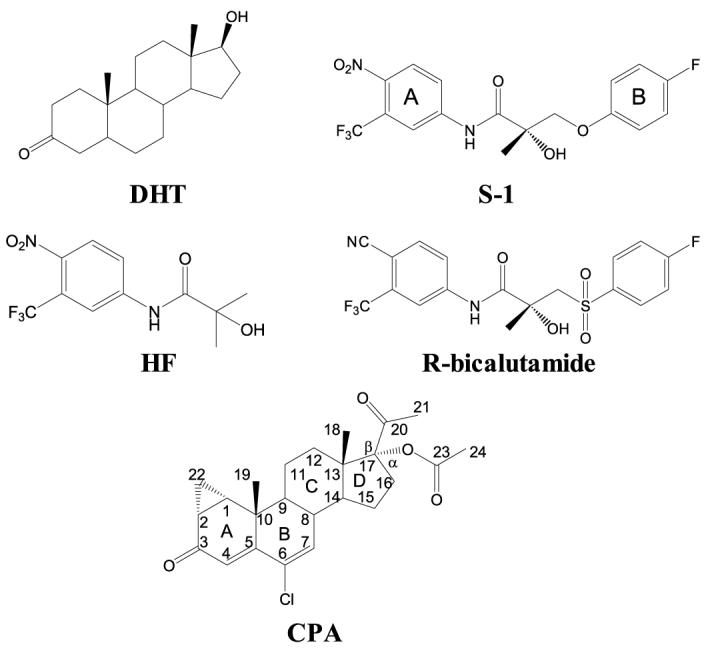
Structures of AR ligands.
Comparison of T877A·CPA and WT·DHT AR LBD
The steroid scaffold of CPA binds in the same plane as DHT (Fig. 2b). The increased bulkiness of CPA in comparison to DHT is a result of a number of structural modifications, including a three-member ring incorporating the 1- and 2-positions of the steroid core structure, a chlorine group at the 6-position, an acetyl group substituted in place of the 17β-hydroxyl group, and an oxygen-linked acetate group also branched from the 17-position. The three-member ring on the A-ring of CPA is accommodated without inducing any changes to AR residues relative to the WT·DHT structure (16, 21). Additionally, the chlorine atom does not cause any remarkable alteration to the positions of AR residues, but the A- and B-rings of CPA are rotated slightly away from Val-746 to alleviate steric contacts. The 17β-acetyl group and 17β-hydroxyl of CPA and DHT, respectively, both form hydrogen bonds to Asn-705. The rotation of the amide on the Asn-705 side chain in the DHT-bound crystal structure is unclear due to the ability of the hydroxyl group to act as both a donor and acceptor and has been reported in both conformations (16, 21). However, the Asn-705 side chain in the CPA-bound AR must be oriented such that the amine is rotated toward the ketone of the acetyl group of CPA for hydrogen bond formation. Thr-877 also forms a hydrogen bond to the 17β-hydroxyl group of DHT, yet this hydrogen bond would likely not occur when CPA is bound to the WT AR as seen from the orientation of the 17β-acetyl group adopted in the T877A AR. Overlay of the WT·DHT and T877A·CPA structures demonstrates that the C-21 methyl group of CPA would be sterically hindered by the presence of the Thr-877 side chain, which may repel helix 11 away from the binding pocket in the WT AR. The acetate group branched from 17α-position of CPA that faces in the opposite direction as the hydroxyl group of DHT is the most significant region of increased bulk on CPA compared with other steroidal ligands for AR. This bulky substituent is accommodated by inducing movement of the Leu-701 side chain (Fig. 2b), which expands the AR binding pocket in this area relative to all AR LBD crystal structures reported to date. More interestingly, the location of Leu-701 in the T877A CPA complex causes the loop between helices 11 and 12 to be drastically repositioned and less ordered than other AR LBD complexes (Fig. 2c). Comparison of the DHT- and CPA-bound crystal structures demonstrates that Leu-701 in the T877A·CPA complex sterically precludes Val-889 from adopting the position seen in the DHT-complexed structure. In turn, this otherwise well ordered loop in the AR LBD is displaced >7.5 Å in some regions, and the C-terminal portion of helix 11 is also unwound.
Transcriptional Activation of CPA in the WT, T877A, and L701A AR
Structural evidence that Leu-701 is displaced by CPA upon AR binding and causes a significant effect on the packing of the loop between helices 11 and 12 led us to the hypothesis that interaction with Leu-701 is important for the antiandrogenic properties of CPA similar to Thr-877. The L701A AR mutant was therefore constructed to investigate whether this mutation would also increase the ability of CPA to stimulate transcriptional activation at low concentrations. In the WT AR, DHT elicited a maximum response by 0.1 nm (Fig. 3a). CPA elicited only a small percentage of transcriptional activation at 100 nm and slightly more at 1000 nm relative to DHT, whereas HF and R-bicalutamide showed nearly no stimulation even at 1000 nm. In the T877A mutant, both CPA and HF elicited a substantial response at 10 nm (Fig. 3b). DHT maintained its potent activity in the T877A AR, and R-bicalutamide showed no response. CPA also induced a drastic increase in transcriptional activation in the L701A AR compared with the WT (Fig. 3c), yet no activity was observed for HF or R-bicalutamide in this mutant. Furthermore, a loss of activity was seen with DHT in the L701A AR inciting a near equal response as CPA in this mutant.
FIGURE 3. Transcriptional activation of DHT, CPA, HF, and R-bicalutamide (R-bic) in the WT (a), T877A (b), and L701A (c) AR variants.
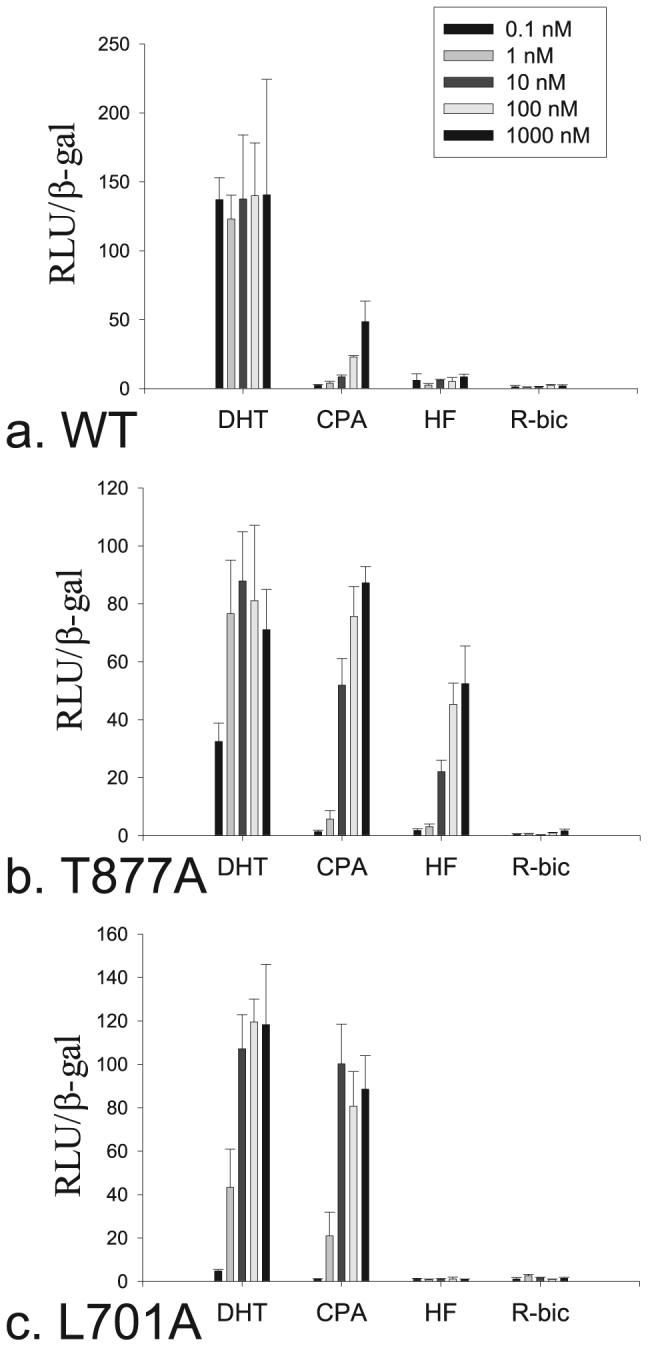
Data are reported as relative luciferase units (RLU) over control normalized by the total amount of β-galactosidase activity in each well.
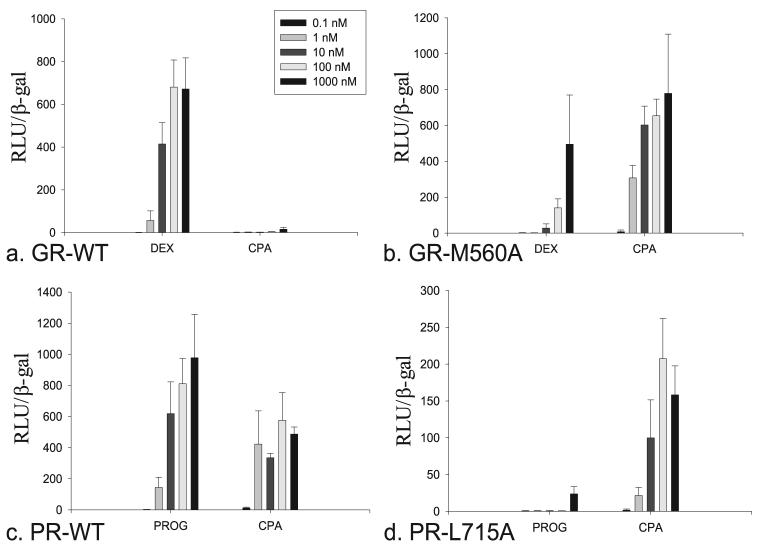
Transcriptional Activation of CPA in the WT and M560A GR
Given the high degree of homology, we aligned the AR with the GR and found that the Met-560 corresponds to the Leu-701 residue (Fig. 2d). Based on the position of the 17α-acetate of CPA in the AR, Met-560 is likely displaced by this substituent upon GR binding thus causing a similar unfolding of the receptor. Therefore, we constructed the M560A mutant in the GR and compared the ability of CPA to induce transcriptional activation relative to the WT receptor. As predicted, this mutation was found to significantly increase ligand-dependent transcription of the GR with CPA. In the WT form of the GR, dexamethasone begins to elicit transcriptional activation at 1 nm with a more prominent effect at 10 nm similar to previous reports (22) (Fig. 4a). Surprisingly, the M560A mutant significantly decreased transcriptional activity nearly 100-fold with dexamethasone (Fig. 4b). Conversely, CPA induces only minute transcriptional activity of the GR in the murine mammary tumor virus reporter assay up to 1000 nm, but the M560A causes CPA to act as a potent agonist for the GR at 1 nm.
FIGURE 4.
Transcriptional activation of dexamethasone and CPA in the WT (a) and M560A (b) GR variants, and progesterone and CPA in the WT (c) and L715A (d) PR variants.
Transcriptional Activation of CPA in the WT and L715A PR
Similarly, we constructed the L715A mutant in the PR, because the Leu-715 corresponds to Leu-701 in the AR. CPA is known to act as an agonist for the WT PR similar to progesterone and has therefore clinical use in hormonal contraception. We found that CPA in fact does elicit transcriptional activation in the murine mammary tumor virus-reporter gene assay at a concentration as low as 1 nm similar to progesterone (Fig. 4c). However, the maximal effect seen with CPA was substantially less than that observed with progesterone. The decrease in activity seen in the L715A mutant with progesterone was extreme with complete loss of transcription up to 100 nm and only a minor effect at 1000 nm. CPA, on the other hand, retained its activity with this mutant (Fig. 4d).
Comparison of Binding Pockets Observed in Various AR LBD Structures
Recently, we reported crystal structures of the AR LBD bound to S-1 in the WT, T877A, and W741L mutants (1, 8). Each of these AR variants accommodated an identical binding conformation of S-1, which included an expansion of the binding pocket as compared with the AR when complexed to steroidal agonists such as DHT and R1881 (16, 23). Although the binding conformation that S-1 and structurally related compounds adopted in the AR was poorly predicted through molecular modeling (11, 13), the x-ray crystal structure of the AR·S-1 complex demonstrated the induced fit of S-1 through displacement of Trp-741 (8). The repositioning of this residue resulted in expansion of the binding pocket (Fig. 5, a and b) for direct ligand interactions with residues in the ligand-dependent activation function (AF-2), including Gln-738 and Ile-898 and a basis for understanding the structure-activity relationships observed with structural analogs of S-1 (15, 24-28). Interestingly, the crystal structure of the T877A·CPA complex demonstrated expansion of the AR binding pocket in the opposite direction as compared with S-1. The induced fit of CPA in the AR from displacement of Leu-701 thus opens another region for which drug interactions can be exploited. In particular, the backbone carbonyl of Ser-778 appears available for hydrogen bonding (Fig. 5c).
FIGURE 5. Expanded binding pocket for CPA in the AR.
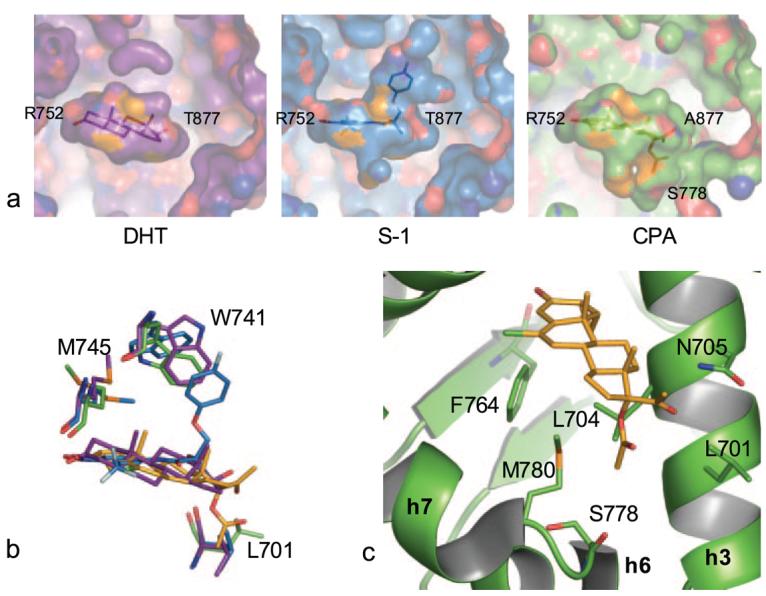
a, comparison of the binding pockets in AR LBD complexes demonstrates the increased binding cavity utilized by S-1 (marine) in comparison to DHT (purple). CPA binds the AR (green) to occupy an additional region located in the opposite direction as the expanded S-1 binding pocket. b, the additional binding regions in the AR are due to the induced fit of S-1 and CPA, which displace Trp-741 and Leu-701, respectively. c, Ser-778 in the distant region of the CPA binding pocket provides a potential hydrogen bond partner that may be exploited in designing new ligands for the AR.
AR Docking Studies
We used AutoDock3.05 to examine docking solutions obtained to various AR LBD crystal structures. DHT was removed and docked into both the WT (PDB code 1I37) and T877A (PDB code 1I38) AR LBD with low r.m.s.d. values from the bound conformation of DHT observed in the crystal structures. Hydrogen bonds to Arg-752, Asn-705, and Thr-877 in the WT and Arg-752 and Asn-705 in the T877A mutant were also present with DHT, further validating the consistency of our docking methods with crystallographic data. In addition, these solutions exhibited low free binding energies (Table 2) with an appropriate increase observed in the T877A docking run due to loss of this hydrogen bond partner. Conversely, the binding energies obtained from docking CPA to the WT AR (PDB code 1I37) and T877A (PDB code 1I38) structures that were co-crystallized with DHT (16) were much higher likely due to the poor ability of these structures to accommodate CPA. The solution with the most favorable free energy of binding showed conservation of the hydrogen bonds to Arg-752 and Asn-705 with the 3-keto group and 17β-acetyl group, respectively, but CPA was rotated out of the steroidal plane relative to the x-ray structure to accommodate the acetate between Leu-701 and Phe-876. The docking solution with the lowest free energy of binding to both the WT and T877A coordinate files demonstrated the same conformation for CPA, yet a substantial improvement in binding energy to the T877A structure was observed likely due to the lack of a steric interaction with the Thr-877 side chain. CPA was docked to the T877A AR structure reported herein with a significantly lower predicted binding energy as compared with the DHT-complexed structures, and an orientation almost identical to that observed in the x-ray crystal structure as demonstrated by the r.m.s.d. of only 0.45 Å. The T877A·CPA structure is thus useful for predicting binding conformations of ligands that occupy this expanded cavity and studying interactions in this otherwise occluded region of the AR in silico.
TABLE 2.
Docking results
| Ligand | Receptor (PDB Code) | Binding energy | r.m.s.d.a (relative to x-ray) |
|---|---|---|---|
| kcal/mol | Å | ||
| DHT | WT-DHT (1I37) | −12.74 | 0.36 |
| DHT | T877A-DHT (1I38) | −12.22 | 0.67 |
| CPA | WT-DHT (1I37) | −8.75 | 1.32 |
| CPA | T877A-DHT (1I38) | −9.92 | 1.32 |
| CPA | T877A-CPA (2Oz7) | −11.38 | 0.45 |
r.m.s.d. of the docked ligand from the conformation of the ligand observed in the x-ray crystal structure.
DISCUSSION
Herein we presented the crystal structure of CPA bound to the T877A AR LBD. Based on the structural findings observed in the T877A·CPA complex, we predicted that the L701A AR mutant would increase agonist activity of CPA due to the relief of the strain on helix 3 and recovery of the packing of the loop between helices 11 and 12 into the groove observed in other AR LBD structures. As predicted, CPA stimulated transcriptional activation in the L701A AR similar to T877A AR. Conversely, HF exhibited no response in the L701A AR, demonstrating the ability of this mutation to selectively confer agonist activity to CPA. This result further suggests that the gain in AR function observed in the L701A AR is due to the interaction with CPA, because HF does not alter the position of Leu-701 in the T877A AR LBD (8) relative to the published WT·DHT structures (16, 21). Electron density maps for AR residues 886–890 in the T877A·CPA structure demonstrated that this region is not well ordered. The flexibility of this loop, however, provides insight to understanding the dynamics of the AR LBD. Unlike other published AR LBD complexes, the T877A·CPA structure elucidates a gateway for the ligand to enter the binding pocket. Crystal structures of the estrogen receptor α LBD have shown that the dynamic properties of helix 12 act as a lid that provides an entrance for ligands to reach the binding cavity (9). However, the C terminus of the AR contains a β-sheet that may restrict the mobility of helix 12 relative to the crystal structures of the estrogen receptor α, which were determined with a truncated C terminus. Furthermore, evidence of the alteration in the folding of C terminus of helix 11 and adjacent loop region in the T877A·CPA structure suggests that this dynamic portion of the AR may provide ligand accessibility to the binding pocket. As seen from Fig. 2 (b and c), it can be anticipated that CPA binding in the presence of the Thr-877 side chain would push helix 11 away from the binding pocket and thus intensify the unfolding of the AR LBD that already occurs in this region. Such an alteration to the AR LBD structure would resemble the change induced by the ligand, GC-24, in the human thyroid hormone receptor β (29). Transcriptional activation data suggest that both the T877A and L701A AR relieve a sufficient amount of strain when complexed to CPA to recover functional AF-2 formation as seen from the ability of CPA to induce transcriptional activation at a low concentration (i.e. 10 nm). Conversely, bicalutamide likely unfolds the AR LBD at the N terminus of helix 12 from steric interaction with Met-895 as shown from our previous report in which W741L and M895T convert it to an agonist by reducing the bulk in this region of the AR (8).
Although CPA acts as an antagonist in the GR similar to the AR, it is an agonist ligand for the PR. The corresponding residue to Leu-701 in the GR and PR were mutated to decrease bulk in the region presumably occupied by the 17α-acetate of CPA upon binding to these other steroid receptors. The M560A mutant in the GR predictably converted CPA from an antagonist in the WT (22) into a potent agonist. This result is explained by the ability of the GR to accommodate the 17α-acetate of CPA and prevent disruption of the GR-LBD folding of the loop linking helices 11 and 12. The M560A mutant severely weakened the activity of dexamethasone, which is likely attributed to loss of van der Waals contacts. Both CPA and progesterone displayed potent activity in the WT PR eliciting transcriptional activation at low nanomolar concentrations as expected. However, almost complete loss of activity was observed with progesterone in the L715A mutant with preservation of CPA activity.
The expanded binding pocket seen in the AR T877A·CPA complex demonstrates an expanded binding pocket not seen in other reported AR LBD crystal structures to date. This finding explains the discrepancy observed in docking experiments and the incompatibility of CPA with the ligand-binding pocket in the AR from the WT·DHT and T877A·DHT complexes (16). The receptor interactions in this cavity provide information useful for structure-based drug design. Ligands that hydrogen bond to Ser-778 will likely offer a means of optimizing binding affinities of new compounds that target the AR. Unfortunately, receptor flexibility is difficult to predict with molecular modeling. Docking software such as FlexX and AutoDock employ rigid protein structures, which limits the ability to accurately predict binding conformations of AR ligands (10, 15). Here we demonstrate the use of ligand-receptor co-crystallography to determine flexible regions of the AR and identify new insight for structure-based drug design and receptor function.
Footnotes
This work was supported by National Institutes of Health Grants R01 DK59800 and R01 DK065227.
The atomic coordinates and structure factors (code 2Oz7) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
The abbreviations used are: AR, androgen receptor; CPA, cyproterone acetate; DHT, dihydrotestosterone; GR, estrogen receptor; HF, hydroxyflutamide; LBD, ligand-binding domain; PR, progesterone receptor; r.m.s.d., root mean square distance; S-1, S-3-(4-fluorophenoxy)-2-hydroxy-2-methyl-N-[4-nitro-3-(trifluoromethyl)phenyl] propionamide; GST, glutathione S-transferase; DTT, dithiothreitol; CHAPS, 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonic acid; WT, wild type; CMV, cytomegalovirus.
W. L. DeLano (2002) The PyMOL Molecular Graphics System, DeLano Scientific, San Carlos, CA.
REFERENCES
- 1.Bohl CE, Gao W, Miller DD, Bell CE, Dalton JT. Proc. Natl. Acad. Sci. U. S. A. 2005;102:6201–6206. doi: 10.1073/pnas.0500381102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yoshida T, Kinoshita H, Segawa T, Nakamura E, Inoue T, Shimizu Y, Kamoto T, Ogawa O. Cancer Res. 2005;65:9611–9616. doi: 10.1158/0008-5472.CAN-05-0817. [DOI] [PubMed] [Google Scholar]
- 3.Veldscholte J, Berrevoets CA, Ris-Stalpers C, Kuiper GG, Jenster G, Trapman J, Brinkmann AO, Mulder E. J. Steroid Biochem. Mol. Biol. 1992;41:665–669. doi: 10.1016/0960-0760(92)90401-4. [DOI] [PubMed] [Google Scholar]
- 4.Poujol N, Wurtz JM, Tahiri B, Lumbroso S, Nicolas JC, Moras D, Sultan C. J. Biol. Chem. 2000;275:24022–24031. doi: 10.1074/jbc.M001999200. [DOI] [PubMed] [Google Scholar]
- 5.Tan J, Sharief Y, Hamil KG, Gregory CW, Zang DY, Sar M, Gumerlock PH, DeVere White RW, Pretlow TG, Harris SE, Wilson EM, Mohler JL, French FS. Mol. Endocrinol. 1997;11:450–459. doi: 10.1210/mend.11.4.9906. [DOI] [PubMed] [Google Scholar]
- 6.Suzuki H, Akakura K, Komiya A, Aida S, Akimoto S, Shimazaki J. Prostate. 1996;29:153–158. doi: 10.1002/1097-0045(199609)29:3<153::aid-pros2990290303>3.0.co;2-5. [DOI] [PubMed] [Google Scholar]
- 7.Hara T, Miyazaki J, Araki H, Yamaoka M, Kanzaki N, Kusaka M, Miyamoto M. Cancer Res. 2003;63:149–153. [PubMed] [Google Scholar]
- 8.Bohl CE, Miller DD, Chen J, Bell CE, Dalton JT. J. Biol. Chem. 2005;280:37747–37754. doi: 10.1074/jbc.M507464200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brzozowski AM, Pike AC, Dauter Z, Hubbard RE, Bonn T, Engstrom O, Ohman L, Greene GL, Gustafsson JA, Carlquist M. Nature. 1997;389:753–758. doi: 10.1038/39645. [DOI] [PubMed] [Google Scholar]
- 10.Marhefka CA, Moore BM, 2nd, Bishop TC, Kirkovsky L, Mukherjee A, Dalton JT, Miller DD. J. Med. Chem. 2001;44:1729–1740. doi: 10.1021/jm0005353. [DOI] [PubMed] [Google Scholar]
- 11.Bohl CE, Chang C, Mohler ML, Chen J, Miller DD, Swaan PW, Dalton JT. J. Med. Chem. 2004;47:3765–3776. doi: 10.1021/JM0499007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Balog A, Salvati ME, Shan W, Mathur A, Leith LW, Wei DD, Attar RM, Geng J, Rizzo CA, Wang C, Krystek SR, Tokarski JS, Hunt JT, Gottardis M, Weinmann R. Bioorg Med. Chem. Lett. 2004;14:6107–6111. doi: 10.1016/j.bmcl.2004.09.049. [DOI] [PubMed] [Google Scholar]
- 13.Soderholm AA, Lehtovuori PT, Nyronen TH. J. Med. Chem. 2005;48:917–925. doi: 10.1021/jm0495879. [DOI] [PubMed] [Google Scholar]
- 14.Lill MA, Winiger F, Vedani A, Ernst B. J. Med. Chem. 2005;48:5666–5674. doi: 10.1021/jm050403f. [DOI] [PubMed] [Google Scholar]
- 15.Marhefka CA, Gao W, Chung K, Kim J, He Y, Yin D, Bohl C, Dalton JT, Miller DD. J. Med. Chem. 2004;47:993–998. doi: 10.1021/jm030336u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sack JS, Kish KF, Wang C, Attar RM, Kiefer SE, An Y, Wu GY, Scheffler JE, Salvati ME, Krystek SR, Jr., Weinmann R, Einspahr HM. Proc. Natl. Acad. Sci. U. S. A. 2001;98:4904–4909. doi: 10.1073/pnas.081565498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kauppi B, Jakob C, Farnegardh M, Yang J, Ahola H, Alarcon M, Calles K, Engstrom O, Harlan J, Muchmore S, Ramqvist AK, Thorell S, Ohman L, Greer J, Gustafsson JA, Carlstedt-Duke J, Carlquist M. J. Biol. Chem. 2003;278:22748–22754. doi: 10.1074/jbc.M212711200. [DOI] [PubMed] [Google Scholar]
- 18.Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL. Acta Crystallogr. D. Biol. Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 19.Jones TA, Zou JY, Cowan SW, Kjeldgaard Acta Crystallogr. Sect. A. 1991;47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 20.Goodsell DS, Morris GM, Olson AJ. J. Mol. Recognit. 1996;9:1–5. doi: 10.1002/(sici)1099-1352(199601)9:1<1::aid-jmr241>3.0.co;2-6. [DOI] [PubMed] [Google Scholar]
- 21.Hur E, Pfaff SJ, Payne ES, Gron H, Buehrer BM, Fletterick RJ. PLoS. Biol. 2004;2:E274. doi: 10.1371/journal.pbio.0020274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Honer C, Nam K, Fink C, Marshall P, Ksander G, Chatelain RE, Cornell W, Steele R, Schweitzer R, Schumacher C. Mol. Pharmacol. 2003;63:1012–1020. doi: 10.1124/mol.63.5.1012. [DOI] [PubMed] [Google Scholar]
- 23.Matias PM, Donner P, Coelho R, Thomaz M, Peixoto C, Macedo S, Otto N, Joschko S, Scholz P, Wegg A, Basler S, Schafer M, Egner U, Carrondo MA. J. Biol. Chem. 2000;275:26164–26171. doi: 10.1074/jbc.M004571200. [DOI] [PubMed] [Google Scholar]
- 24.Kim J, Wu D, Hwang DJ, Miller DD, Dalton JT. J. Pharmacol. Exp. Ther. 2005;315:230–239. doi: 10.1124/jpet.105.088344. [DOI] [PubMed] [Google Scholar]
- 25.Yin D, He Y, Perera MA, Hong SS, Marhefka C, Stourman N, Kirkovsky L, Miller DD, Dalton JT. Mol. Pharmacol. 2003;63:211–223. doi: 10.1124/mol.63.1.211. [DOI] [PubMed] [Google Scholar]
- 26.Chen J, Hwang DJ, Chung K, Bohl CE, Fisher SJ, Miller DD, Dalton JT. Endocrinology. 2005;146:5444–5454. doi: 10.1210/en.2005-0732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dalton JT, Mukherjee A, Zhu Z, Kirkovsky L, Miller DD. Biochem. Biophys. Res. Commun. 1998;244:1–4. doi: 10.1006/bbrc.1998.8209. [DOI] [PubMed] [Google Scholar]
- 28.He Y, Yin D, Perera M, Kirkovsky L, Stourman N, Li W, Dalton JT, Miller DD. Eur. J. Med. Chem. 2002;37:619–634. doi: 10.1016/s0223-5234(02)01335-1. [DOI] [PubMed] [Google Scholar]
- 29.Borngraeber S, Budny MJ, Chiellini G, Cunha-Lima ST, Togashi M, Webb P, Baxter JD, Scanlan TS, Fletterick RJ. Proc. Natl. Acad. Sci. U. S. A. 2003;100:15358–15363. doi: 10.1073/pnas.2136689100. [DOI] [PMC free article] [PubMed] [Google Scholar]