Abstract
Tyrosyl-DNA phosphodiesterase 1 (Tdp1) cleaves the phosphodiester bond between a covalently stalled topoisomerase I (Topo I) and the 3′ end of DNA. Stalling of Topo I at DNA strand breaks is induced by endogenous DNA damage and the Topo I-specific anticancer drug camptothecin (CPT). The H493R mutation of Tdp1 causes the neurodegenerative disorder spinocerebellar ataxia with axonal neuropathy (SCAN1). Contrary to the hypothesis that SCAN1 arises from catalytically inactive Tdp1, Tdp1−/− mice are indistinguishable from wild-type mice, physically, histologically, behaviorally, and electrophysiologically. However, compared to wild-type mice, Tdp1−/− mice are hypersensitive to CPT and bleomycin but not to etoposide. Consistent with earlier in vitro studies, we show that the H493R Tdp1 mutant protein retains residual activity and becomes covalently trapped on the DNA after CPT treatment of SCAN1 cells. This result provides a direct demonstration that Tdp1 repairs Topo I covalent lesions in vivo and suggests that SCAN1 arises from the recessive neomorphic mutation H493R. This is a novel mechanism for disease since neomorphic mutations are generally dominant.
Keywords: camptothecin, neurodegeneration, SCAN1, Tdp1, topoisomerase I
Introduction
DNA topoisomerases, glycosylases, methyltransferases, and recombinases act via formation of a transient covalent intermediate with DNA. When these DNA-processing enzymes become covalently trapped on the DNA, they cause a particularly harmful kind of DNA damage. The repair pathways for these types of lesions are of great interest because they influence the effectiveness of widely used antibacterial and antitumor drugs that act by stabilizing such covalent complexes (Connelly and Leach, 2004).
Inherited defects of DNA repair are associated with a predisposition to cancer and neurological abnormalities (Friedberg et al, 2006). To our knowledge, spinocerebellar ataxia with axonal neuropathy (SCAN1) is the first example of a human genetic disorder that results from a failure to repair DNA–protein covalent complexes. More importantly, the mutant protein responsible for the disease becomes itself covalently trapped on the DNA.
SCAN1 is an autosomal recessive disorder characterized by ataxia, cerebellar atrophy, and peripheral neuropathy (Takashima et al, 2002). The patients are usually wheelchair bound by early adulthood but retain normal cognitive function suggesting that the disease arises from degeneration or impairment of specific neurons (Takashima et al, 2002). SCAN1 has been associated with the TDP1 1478A>G mutation, which encodes the missense change H493R that disrupts the active site of tyrosyl-DNA phosphodiesterase 1 (Tdp1) (Interthal et al, 2001, 2005b; Takashima et al, 2002).
Tdp1 catalyzes the hydrolysis of the phosphodiester bond between a DNA 3′ end and a tyrosine residue, a linkage specific to the enzyme–DNA covalent complex formed when a type IB DNA topoisomerase cleaves DNA (Yang et al, 1996). Topoisomerase I (Topo I) becomes covalently trapped in a dead-end complex on the DNA when it fails to religate the DNA after cleavage near endogenous lesions (nicks, gaps, or abasic sites) (Pommier, 2004). Tdp1 participates in the repair of Topo I–DNA complexes as a member of the mammalian DNA single-strand break repair complex (Pouliot et al, 2001; El-Khamisy et al, 2005; Interthal et al, 2005b).
The anticancer drug CPT also causes Topo I to stall on the DNA (Pourquier and Pommier, 2001). The predominant cytotoxic lesion is a DNA double-strand break resulting from the collision of a replication fork with the covalent Topo I–DNA complex (D'Arpa et al, 1990). In yeast, these DNA double-strand breaks can also be repaired through Tdp1-independent pathways involving the structure-specific endonucleases Rad1/Rad10 and Mus81/Mms4 (Liu et al, 2002; Vance and Wilson, 2002).
Consistent with these proposed functions of Tdp1, the lymphoblastoid cell lines from SCAN1 patients are CPT hypersensitive (Interthal et al, 2005b) and accumulate CPT-induced DNA single- and double-strand breaks (El-Khamisy et al, 2005; El-Khamisy and Caldecott, 2007). Furthermore, Miao et al (2006) recently showed that SCAN1 cells accumulate Topo I–DNA complexes. These observations have led to the conclusion that SCAN1 arises solely from the catalytic inactivity of H493R Tdp1 (El-Khamisy et al, 2005).
Despite the role of Tdp1 in DNA repair, SCAN1 patients do not have an increased incidence of cancer or other health problems (Takashima et al, 2002). Nor do cancer patients receiving Topo I inhibitors such as CPT have an increased incidence of neurodegeneration. These observations suggest that the etiology of SCAN1 is more complex than simple loss of Tdp1 function.
We describe the first mouse model to study the etiology of SCAN1 and the organismal function of Tdp1. Tdp1-deficient mice are CPT and bleomycin hypersensitive, but do not develop SCAN1-like symptoms. However upon CPT treatment, Tdp1-deficient mouse cells expressing H493R Tdp1 accumulate DNA breaks to higher levels than control Tdp1−/− cells. These data suggest that in addition to reduced Tdp1 activity, the qualitative change in enzymatic activity of human H493R Tdp1 that causes it to become covalently trapped on the DNA may contribute to the etiology of SCAN1.
Results and discussion
Generation and phenotypic analysis of Tdp1−/− mice
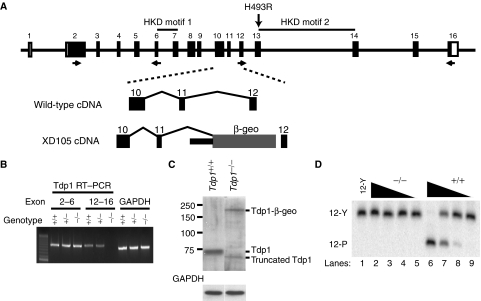
To ascertain whether SCAN1 arises from loss of functional Tdp1, we generated Tdp1-deficient mice (hereafter referred to as Tdp1−/− mice) using a Geo trap inserted in Tdp1 intron 11 (Figure 1A). Embryonic fibroblasts and neurospheres derived from the Tdp1−/− mice did not express full-length Tdp1 mRNA and protein (Figure 1B, C) but did express a stable truncated protein product and a geoTDP1 protein derivative (Figure 1C). The truncated protein was translated from an aberrantly spliced mRNA that encoded a protein without an active site (data not shown), and consistent with this, Tdp1 enzymatic activity was undetectable (Figure 1D). On a 129Sv genetic background, Tdp1+/− parents produced all genotypic classes in Mendelian ratios. Thus, in contrast to Drosophila (Dunlop et al, 2004) in which deficiency of glaikit, the Tdp1 homolog, causes defective embryonic neurogenesis and embryonic lethality, Tdp1-deficient mice did not exhibit embryonic or neonatal lethality.
Figure 1.
Generation of the Tdp1−/− mouse by insertional mutagenesis. (A) Diagram of the mouse Tdp1 genomic structure (top). The H493R mutation found in SCAN1 patients would be encoded in exon 13 of mouse Tdp1. For generation of the Tdp1-deficient mice (Tdp1−/−), we used embryonic stem cells containing a pGT1Lxf insertion in Tdp1 intron 11. This insertion truncates the Tdp1 mRNA following exon 11. The location of the primers used for genotyping and RT–PCR detection are indicated. (B) RT–PCR confirmation of the absence of full-length Tdp1 mRNA expression in the brain of Tdp1−/− mice. The upstream RT–PCR primers reside in exons 2 and 6, the downstream primers in exons 12 and 16. (C) Western confirmation of the absence of expression of full-length Tdp1 protein in Tdp1−/− neurospheres. The western analysis also detected the Tdp1-β-geo fusion protein and a smaller Tdp1 product that is the product of alternative splicing deleting exons 12 and 13. (D) Absence of Tdp1 enzymatic activity in lysates from Tdp1−/− mouse neurospheres as shown by the failure to hydrolyze the tyrosyl-DNA substrate 12-Y. Each lane represents a serial 10-fold dilution of the Tdp1−/− (lanes 2–5) and Tdp1+/+ (lanes 6–9) cell extracts.
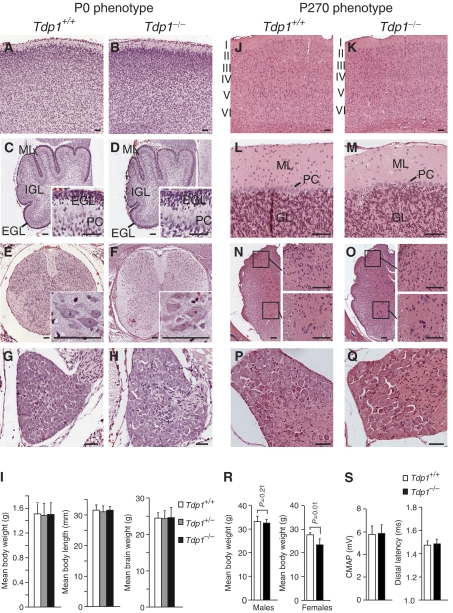
At birth, the Tdp1−/− mice had body weights and lengths, brain weights and histopathology comparable to Tdp1+/+ mice (Figure 2A–I). This phenotype correlates with that of SCAN1 patients who had normal growth and motor, language, social and intellectual development throughout childhood (Takashima et al, 2002). However, in contrast to the SCAN1 patients who develop clinical symptoms in their second decade of life (Takashima et al, 2002), the Tdp1−/− mice examined at 270 days by histopathology and peripheral nerve electrophysiology did not show abnormalities (Figure 2J–S) and those analyzed at 14–17 months did not exhibit behavioral or other abnormalities. Thus, we propose that the Tdp1−/− mice are able to repair endogenous levels of Topo I–DNA complexes by alternate DNA-repair pathways.
Figure 2.
Comparative phenotypic and histopathological evaluation of the Tdp1+/+ and Tdp1−/− mice at P0 (A–I) and P270 (J–S). P0 mice: Hematoxylin and eosin (H&E) staining of cortex (A, B), cerebellar vermis (C, D), spinal cord (E, F), and dorsal root ganglion (G, H). Note the comparability of the external granular cell (EGL), molecular (ML), and internal granular cell layers (IGL) as well as the Purkinje cell number (PC). (I) Comparison of the body weights and lengths and brain weights of Tdp1+/+, Tdp1+/−, and Tdp1−/− littermates. P270 mice: H&E staining of cortex (J, K), cerebellar vermis (L, M), spinal cord (N, O), and dorsal root ganglia (P, Q). The cortical layers are labeled in panels J and K and the cerebellar molecular (ML) and granular cell (GL) layers in panels L and M. The insets in panels N and O are higher magnifications of the dorsal (upper) and ventral (lower) horns. (R) Comparison of male and female body weights. (S) Comparison of the peripheral nerve electrophysiology of Tdp1+/+ and Tdp1−/− littermates. Scale bar=50 μm.
Alternative pathways for the repair of Topo I–DNA covalent complexes
In Saccharomyces cerevisiae, the two best-studied alternative pathways for repairing stalled Topo I are those mediated by the structure-specific endonucleases Rad1/Rad10 and Mus81/Mms4 (Liu et al, 2002; Vance and Wilson, 2002), which correspond to mammalian XPF/ERCC1 and Mus81/Eme1, respectively (Matsunaga et al, 1995; Brookman et al, 1996; Ciccia et al, 2003; Ogrunc and Sancar, 2003). By in situ hybridization and RT–PCR across a spectrum of Tdp1+/+ mouse tissues, Xpf, Ercc1, and Mus81 were expressed at each time point investigated, whereas Eme1 generally had much lower neural expression except in the developing cerebellum (Supplementary Figures 1 and 2). Notably, Eme1 was expressed in non-neural tissues such as the thymus, heart, intestine, liver, skin, vertebrae, teeth, and testes (Supplementary Figure 1M, N). We observed a similar lack of neural expression of Eme1 in the adult human brain by RT–PCR (Supplementary Figure 2T) and northern analyses (data not shown). The mRNAs encoding these endonucleases (Supplementary Figure 2U) as well as the mRNAs encoding the base excision repair enzymes Apex1 and Apex2 (data not shown) were not increased in Tdp1−/− brain tissue. Thus, although not upregulated in Tdp1−/− tissue, the XPF/ERCC1 pathway may function as one alternative pathway for repair of Topo I–DNA complexes in the absence of Tdp1 (Vance and Wilson, 2002), and the low or absent Eme1 expression in the majority of neuronal tissues might contribute to the neuronal specificity of SCAN1.
Tdp1 is expressed in the nervous system of mice and humans
To test whether variation in expression across species accounted for the difference in phenotype between Tdp1−/− mice and SCAN1 patients, we compared the expression of Tdp1 in mice and humans. By northern blot analysis, human Tdp1 mRNA was present in all analyzed adult tissues (Supplementary Figure 3A) and in all tested areas of the central nervous system (Supplementary Figure 3B). Similarly, mouse Tdp1 mRNA was expressed in all analyzed adult tissues (Supplementary Figure 3C) and in the brain from embryonic day (E) 9.5 through adulthood (Supplementary Figure 3D).
In the human brain, Tdp1 was highly expressed in neurons from 10 gestational weeks (GW) through 16 years of age and at very low levels in glia (Figure 3A–C). Similar to the Drosophila glaikit protein (Dunlop et al, 2000), human Tdp1 was expressed in subependymal neural progenitors and cultured neurospheres (Figure 3A and Supplementary Figure 4). Cerebellar, granule and Purkinje cells (Figure 3D–F) and neurons of the dentate nucleus (Figure 3G), spinal cord (Figure 3H), and dorsal root ganglia (DRG) (Figure 3I), the neurons putatively affected by SCAN1 (Takashima et al, 2002), also expressed Tdp1. Similar to the glaikit protein in Drosophila, the Tdp1 protein was prominently expressed in the cytoplasm of some neurons (Figure 3F–H), a poorly understood finding; however, in contrast to human Tdp1, glaikit is not thought to participate in DNA repair but in localization of membrane proteins (Dunlop et al, 2000, 2004). Antiserum nonspecificity does not account for this staining pattern because we obtained the same results with two independently produced antisera, and the preimmune serum did not recognize either a nuclear or a cytoplasmic antigen (Figure 3J–K). Moreover, the antisera recognized a 70 kDa protein in mouse Tdp1−/− fibroblasts transfected with a human Tdp1 expression plasmid but not in untransfected Tdp1−/− fibroblasts (Figure 3L), and competitive inhibition with recombinant Tdp1 protein blocked interaction of the anti-human (Figure 3M) and anti-mouse sera with Tdp1 (data not shown). Although we observed this staining pattern in the tissues from several individuals, this finding could be an artefact of tissue fixation since in our experience Tdp1 readily diffuses out of the nucleus of dying human and mouse cells. Alternatively, this might suggest either cytoplasmic sequestration or a cytoplasmic function for Tdp1 in humans but not in mice.
Figure 3.
Immunohistochemical analysis of Tdp1 protein expression in the human brain. (A) Tdp1 expression in the cerebrum of a human 10 gestational week (GW) brain. Cells adjoining the ventricle (V), and in the subventricular zone (SVZ), intermediate zone (IMZ), and cortical plate (CP) express Tdp1. (B, C) Tdp1 expression in layer two of the cortex from a 40 GW and a 16-year brain. (D–F) Tdp1 expression in the cerebella from a 22 GW, a 9-month and a 4-year brain. Note the staining in the external granular cell layer (EGL), Purkinje cells (PC), and internal granular cell layer (IGL). (G) Tdp1 expression in a 19-year dentate gyrus. (H, I) Tdp1 expression in spinal cord anterior horn cells (AHC) and a dorsal root ganglion of a 10-year old, respectively. (J–M) Specificity of the immune serum is shown by absence of staining of the spinal cord (J) and dorsal root ganglion (K) from a 10-year old with preimmune serum and also by absence of antigen recognition in mouse Tdp1−/− mouse embryonic fibroblasts compared to Tdp1−/− fibroblasts transfected with a human Tdp1 expression construct (Tdp1−/−+pCMV5·Tdp1) (L), and competitive inhibition of the antiserum by recombinant human Tdp1 (M). Scale bar=50 μm.
With the exception of the cytoplasmic localization, the mouse brain showed a similar expression pattern for the Tdp1 RNA and protein (Figure 4). This suggested to us that SCAN1 arises either from loss of a human-specific function as reflected by the differences in protein localization or from a novel function of Tdp1 created by the H493R mutation.
Figure 4.
In situ hybridization and immunohistochemistry showing the spatial and temporal expression of mouse Tdp1 mRNA and protein, respectively. Localization of the Tdp1 mRNA in P1 (A–D), P10 (E–H), and adult (I–L) mice. (A, E, I) Expression of Tdp1 mRNA in a midline sagittal section from a P1, P10, and adult mouse or brain, respectively. Note that Tdp1 is expressed throughout the cortex (Cx), cerebellum (Cb), spinal cord (SC), and dorsal root ganglia (DRG) as well as most other organs including the skin, thymus (Th), heart, lungs, liver, and intestines. (B, F, J) Expression of Tdp1 mRNA in the cortex (Cx) and hippocampus (Hip) of a P1, P10, and adult brain, respectively. (C, G, K) Expression of Tdp1 mRNA in a coronal section of a P1, P10, and adult cervical spinal cord, respectively. The highest expression is in the dorsal horns (DH) and the ventral horns (VH). (D, H, L) Expression of Tdp1 mRNA in a cross-section of a P1, P10, and adult dorsal root ganglion, respectively. (M–O) Serial sections from the P1 mouse hybridized with a sense probe for Tdp1 showing the specificity of the hybridization for the antisense probe used in panels A–L. Scale bar=1 cm (A, E, I, M), 0.5 mm (B, C, F, G, J, K, N, O), 0.2 mm (D, H, L). Localization of the Tdp1 protein in the P0 and the P270 CNS (P-AA). (P) P0 cortex and hippocampus, (Q) P0 layer II cortical neurons, (R) P270 cortex, (S) P270 layer II cortical neurons, (T) P0 cerebellum, (U) P270 cerebellum, (V) P270 dentate nucleus, (W) P270 spinal cord. Nonspecific staining of the P0 cortex (X), P270 cortex (Y), P0 cerebellum (Z), and P270 cerebellum (AA). Scale bar=0.5 mm (P, R, T, U, Y, Z, AA, BB), 0.2 mm (X), 50 μm (Q, S, and all insets). Abbreviations: EGL, external granular layer; PC, Purkinje cell; ML, molecular layer; VH, ventral horn; VHC, ventral horn cell; DH, dorsal horn.
Tdp1−/− mice, neurospheres, and embryonic fibroblasts are CPT hypersensitive
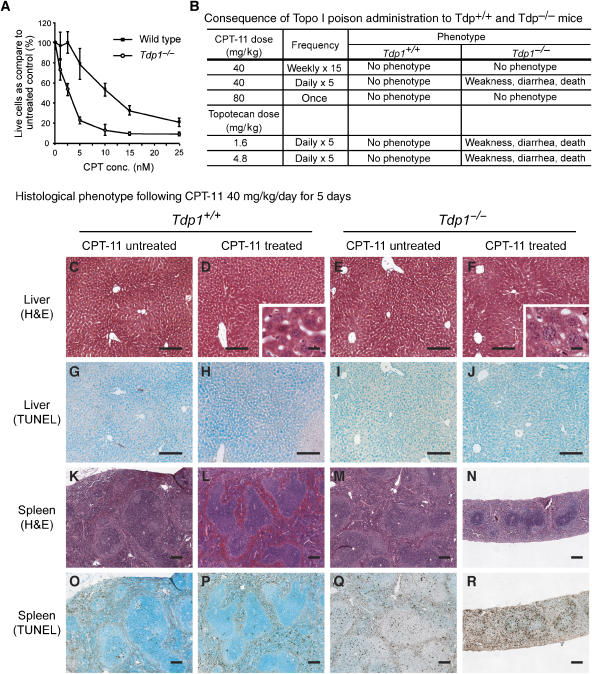
Like cells from the SCAN1 patients (El-Khamisy et al, 2005; Interthal et al, 2005b), Tdp1−/− mouse embryonic fibroblasts (MEFs, Supplementary Figure 6A, D–G) and neurospheres (Figure 5A) were hypersensitive to CPT. Therefore, we hypothesized that the Tdp1−/− mice, and perhaps in particular the nervous system of these mice, would have increased sensitivity to CPT and that CPT treatment might induce a SCAN1-like phenotype. Intraperitoneal administration of the Topo I poison CPT-11 as a single (80 mg/kg) or a weekly (40 mg/kg, for 20 weeks) dose elicited no detectable phenotype in Tdp1−/− or Tdp1+/+ mice aged 70–210 days (Figure 5B). But five consecutive daily doses of CPT-11 (40 mg/kg/day) or of topotecan (1.6 or 4.8 mg/kg/day) were toxic to Tdp1−/− mice ranging in age from 70 to 80 days (Figure 5B). Although the same age Tdp1+/+ mice were unaffected, the Tdp1−/− mice developed weakness and diarrhea and expired within 2 days after completion of CPT-11 or topotecan administration. By histopathology and immunohistochemistry, there was extensive necrosis in the more slowly proliferating intestinal, renal, and hepatic tissues (Figure 5C–J), and extensive apoptosis and marked tissue loss in the more rapidly proliferating lymphoid and hematopoietic tissues (Figure 5K–R). The induction of apoptosis in these rapidly proliferating tissues indicates that Tdp1, as expected, is involved in the repair of Topo I-associated DNA double-strand breaks that are generated when the replication machinery collides with CPT-trapped Topo I. The treated Tdp1−/− mice also had electrophysiological changes, prolonged distal latencies, and reduced compound muscle action potentials (Supplementary Figure 5A), changes typically seen in acute illness but different from those of the SCAN1 patients (Takashima et al, 2002). Whether treated with CPT-11 or topotecan, Tdp1−/− mice did not exhibit ataxia or have detectable atrophy or apoptosis in the cerebrum, cerebellum, spinal cord, DRG, or peripheral nerve by histological, immunohistochemical, TUNEL, or ultrastructural analyses (Supplementary Figure 5B–U). In addition, the Tdp1−/− glia and neurons differentiated from neurospheres (Supplementary Figure 5V, W) or cultured Tdp1−/− cortical neurons (data not shown) failed to exhibit cell death or apoptosis at CPT doses that impaired neurosphere proliferation.
Figure 5.
Analysis of CPT-11-treated and untreated Tdp1+/+ and Tdp1−/− cells and mice. (A) CPT sensitivity of Tdp1+/+ and Tdp1−/− neurosphere cells following 72 h of incubation with camptothecin (CPT) at the indicated concentrations. (B) CPT-11 and topotecan treatment protocols and outcomes for Tdp1+/+ and Tdp1−/− mice. Note that only mice treated repetitively at short intervals developed a phenotype; this suggests a high level of redundancy for the removal of stalled Topo I. (C–R) Histopathology and TUNEL staining of liver and spleen derived from Tdp1+/+ and Tdp1−/− mice treated with 40 mg/kg of CPT-11 for 5 days. Note the extensive vacuolization (F) but paucity of TUNEL-positive cells (J) in the liver of Tdp1−/− mice suggesting necrotic cell death. In contrast, the lymphoid and hematopoietic tissues such as the spleen showed marked loss of tissue (M versus N) with a large number of TUNEL-positive cells (Q versus R) suggesting cell death by apoptosis.
In summary, these results confirm that the CPT hypersensitivity of SCAN1 lymphoblastoid cells is likely arising because of deficient Tdp1 activity. However, they also highlight the possibility that the neuropathology of SCAN1 may not be caused solely by deficient Tdp1 activity because accentuation of the formation of Topo I–DNA complexes with the administration of Topo I inhibitors did not cause SCAN1-like neural dysfunction. Rather, nonproliferating Tdp1−/− neural cells in culture and in vivo did not show acute toxicity to Topo I–DNA complexes, whereas proliferating Tdp1−/− cells in vivo were quite sensitive to the toxicity of Topo I–DNA complexes (Figure 5 and Supplementary Figure 5). The resistance of nonproliferating neural cells to topotecan and CPT-11 cannot be ascribed to the failure of these drugs to penetrate the blood–brain barrier since topotecan penetration of the blood–brain barrier is well established in rodents (El-Gizawy and Hedaya, 1999; Zhuang et al, 2006). In addition, the DRG, which resides outside of the blood–brain barrier (Arvidson, 1979), did not exhibit histopathological changes (Supplementary Figure 5).
The resistance of nonproliferating neural cells to CPT might suggest that alternative pathways are able to repair low levels of Topo I–DNA complexes. The existence of such alternative pathways is supported by the observation of Xpf induction in lung tumors commonly resistant to camptothecin (Sestili et al, 2006) and by the increased levels of homologous recombination in SCAN1 patient lymphoblastoid cells (El-Khamisy et al, 2005). Additionally, we have evidence for the existence of an endonuclease-dependent repair pathway for stalled Topo I (J Leppard, H Interthal, and J Champoux, unpublished).
Tdp1−/− mice and embryonic fibroblasts are bleomycin but not etoposide hypersensitive
SCAN1 cells are also deficient in processing of DNA phosphoglycolate 3′ ends (Inamdar et al, 2002; Zhou et al, 2005). Therefore, we hypothesized that the Tdp1−/− cells and mice would have increased sensitivity to bleomycin. By the comet assay, Tdp1−/− MEFs were hypersensitive to bleomycin (Supplementary Figure 6B), and with intraperitoneal administration of the bleomycin for 10 consecutive days (10 mg/kg/day), Tdp1−/− mice developed weakness and expired within 4 days following treatment. Histopathology and immunohistochemistry showed extensive apoptosis and marked tissue loss in the more rapidly proliferating lymphoid and hematopoietic tissues (Supplementary Figure 6H–K). The treated Tdp1−/− mice did not exhibit ataxia or electrophysiological changes typical of SCAN1 or have detectable atrophy or apoptosis in the cerebrum, cerebellum, spinal cord, or DRG peripheral nerve (Supplementary Figure 6L–O and data not shown).
Tdp1 is also proposed to participate in the repair of DNA double-strand breaks resulting from stalling of Topoisomerase II (Topo II) (Barthelmes et al, 2004; Nitiss et al, 2006). If Tdp1 played a key role in this repair process, then Tdp1−/− cells and mice might have increased sensitivity to etoposide, a Topo II inhibitor (Liu, 1989). However, consistent with the studies of lymphoblastoid cells from SCAN1 patients (Interthal et al, 2005b), neither Tdp1−/− MEFs nor mice showed increased sensitivity to etoposide (Supplementary Figure 6C, P–W). Both Tdp1+/+ and Tdp1−/− mice became ill and died 2 or 3 days after 5 consecutive days of 30 or 40 mg/kg/day of etoposide, and both were equally symptom free 60 days after 5 days of 20 mg/kg/day of etoposide. The Tdp1+/+ and Tdp1−/− mice did not have detectable pathological differences by histological, immunohistochemical, or TUNEL analyses (Supplementary Figure 6P–W and data not shown).
These in vivo and cell culture studies confirm the participation of Tdp1 in the repair of DNA phosphoglycolate 3′ ends but not in the repair of Topo II–DNA complexes. Moreover, the absence of an SCAN1-like phenotype in the treated mice again highlights the possibility that SCAN1 might not arise solely from deficient Tdp1 activity.
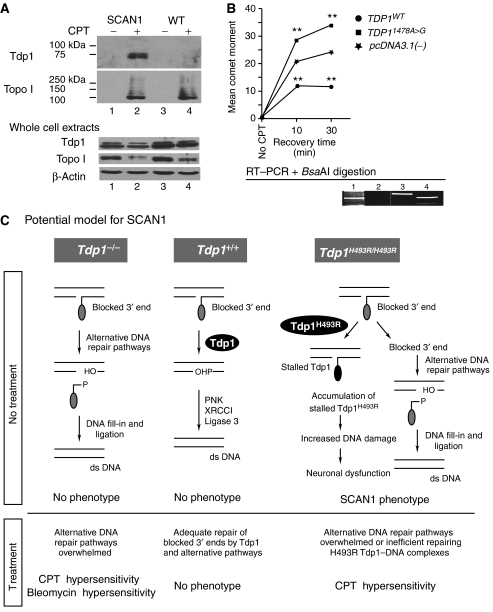
CPT treatment of cells expressing Tdp1 H493R causes the accumulation of H493R–DNA complexes and the accumulation of DNA strand breaks
The above observations suggested that SCAN1 is caused by a novel and distinct function of Tdp1 created by the SCAN1 point mutation. Previously, Interthal et al (2005a, 2005b) hypothesized that SCAN1 might not only arise from the 25-fold reduction in activity caused by the H493R change, but also from the accumulation of the Tdp1 H493R–DNA covalent reaction intermediate which has a half-life of ∼13 min. The removal of H493R Tdp1 from the DNA by wild-type Tdp1 explains the recessive nature of SCAN1 (Interthal et al, 2005a, 2005b). To test whether Tdp1 H493R becomes trapped on the genomic DNA in vivo in response to CPT treatment, we analyzed telomerase-immortalized skin fibroblasts from unaffected controls and SCAN1 patients using a modified in vivo complex of enzyme (ICE) assay (Subramanian et al, 1995). CPT-treated fibroblasts from both sources accumulated the Topo I–DNA covalent intermediate as expected, but only SCAN1 cells accumulated a covalent Tdp1–DNA intermediate (Figure 6A). Based on alkaline comet assay analyses and accumulation of γH2AX, CPT-treated Tdp1−/− MEFs expressing the human H493R Tdp1 protein accumulated DNA strand breaks more rapidly and to a higher level than CPT-treated Tdp1−/− MEFs transfected with the control expression plasmid (Figure 6B and Supplemental Figure 7). In contrast, reduced accumulation of DNA strand breaks was evident upon expressing the wild-type Tdp1 protein (Figure 6B and Supplementary Figure 7).
Figure 6.
Stalling of H493R Tdp1 on DNA and a model for the causation of SCAN1. (A) In vitro complex of enzyme assay showing that treatment of unaffected and SCAN1 patient skin fibroblasts with camptothecin (CPT) causes formation of a covalent Tdp1–DNA complex in the SCAN1 fibroblasts but not in unaffected fibroblasts (upper panel). The expected covalent Topo I–DNA complex (middle panel) and the relative abundance of each protein (lower panel) are also shown. (B) Alkaline comet assay showing increased CPT induction of DNA strand breaks in Tdp1−/− MEFs expressing H493R Tdp1 (pcDNA3.1(−)·TDP11478A>G) and reduced induction of DNA breaks in Tdp1−/− MEFs expressing wild-type Tdp1 (pcDNA3.1(−)·TDP1WT). The comet moments were measured following treatment with 1 μM CPT for 1 h and recovery in CPT-free medium for the indicated times. The comet moment of Tdp1−/− MEFs transfected with only the expression vector (pcDNA3.1(−)) was significantly different from TDP1WT and TDP11478A>G (**P≪0.001). The expression of human wild type and H493R Tdp1 was detected in the transfected cells by western blot (data not shown) and RT–PCR followed by BsaAI digestion of the products (lower panel) as previously described (Takashima et al, 2002). Lane 1: molecular weight markers; lane 2: Tdp1−/− MEFs transfected with plasmid pcDNA3.1(−), the empty vector; lane 3: Tdp1−/− MEFs transfected with plasmid pcDNA3.1(−)·TDP1WT, the expression vector for wild-type human TDP1 cDNA; lane 4: Tdp1−/− MEFs transfected with plasmid pcDNA3.1(−)·TDP11478A>G, the expression vector for human H493R Tdp1 cDNA. Note that the 1478A>G mutation creates a BsaAI restriction site. (C) Potential model for the molecular basis of SCAN1. DNA breaks with blocked 3′ ends (e.g., Topo I or phosphoglycolate) undergo Tdp1-facilitated DNA repair via both DNA single-strand break repair (SSBR) and double-strand break repair (DSBR) mechanisms. With loss of functional Tdp1 (Tdp1−/−), there is sufficient redundant activity for adequate DNA repair by alternative pathways (e.g. endonuclease-dependent pathways) unless the system is further stressed as by administration of CPT or bleomycin. In contrast, when Tdp1 carries the H493R mutation, it not only has a quantitative reduction in overall activity, but also a qualitative change resulting in accumulation of Tdp1–DNA complexes. These complexes are efficiently removed from the DNA by wild-type Tdp1 in all tissues of heterozygotes, whereas they are only removed in replicating cells of homozygotes by alternative DNA strand break repair mechanisms. According to this model, the transcriptional interference and/or apoptosis resulting from the Tdp1–DNA complexes in nondividing neurons causes SCAN1 via neurodegeneration.
These data show that the human H493R Tdp1 has an enzymatic activity in vivo that is qualitatively different from the Tdp1 activity in Tdp1−/− cells or Tdp1−/− cells complemented with wild-type Tdp1. This defines Tdp1 H493R as a neomorphic mutation. Moreover, the covalent stalling of H493R Tdp1 on the DNA in CPT-treated SCAN1 cells provides direct evidence that human Tdp1 removes CPT-trapped Topo I from the DNA in vivo.
Model for the repair of covalent Topo I–DNA complexes and the molecular mechanism of SCAN1
We previously had hypothesized that SCAN1 arises from cell death or cellular malfunction secondary to the accumulation of Topo I–DNA complexes; however, our data here suggest that Tdp1–DNA intermediates arising from antecedent Topo I–DNA complexes may be the underlying basis for the disease (Figure 6C). Although treatment of Tdp1−/− mice with Topo I inhibitors or bleomycin might have recapitulated the SCAN1 phenotype if sufficient DNA damage was able to accumulate, Tdp1−/− mice treated with either high or low doses of drug did not develop an SCAN1-like phenotype. This suggests that the resultant DNA complexes are not the initiating lesions; however, it seems more likely that for the high doses, the drug-induced damage greatly exceeded the repair capacity of the mice and therefore failed to mimic the low level of naturally occurring lesions that accumulate in SCAN1 patients. And for low doses of drug, redundant pathways effectively repaired the DNA damage. Therefore, we hypothesize that the prolonged half-life of the H493R Tdp1–DNA complexes and the increased level of DNA damage in neuronal cells is the origin of the cellular malfunction underlying SCAN1. Such an increase in DNA damage in neuronal cells could arise from decreased efficiency Tdp1–DNA adduct removal since some DNA repair pathways are attenuated in terminally differentiated cells (Nouspikel and Hanawalt, 2002). Testing this model requires analysis of another mouse model solely expressing the mutant H493R Tdp1 rather than the loss of function mutation described here.
Relevant to the treatment of cancer, the absence of detectable acute effects of CPT and bleomycin treatment on the murine nervous system suggests that nonproliferating cells of the nervous system are insensitive to Topo I–DNA complexes and 3′ phosphoglycolate-DNA damage. Thus, in contrast to the accentuation of demyelinating neuropathies by some chemotherapies (Weimer and Podwall, 2006), short-term administration of these chemotherapeutic agents is unlikely to induce neurological disease even in the absence of Tdp1. Additionally, our observations suggest that Tdp1 is also a reasonable chemotherapeutic target as long as the formation and consequences of Tdp1–DNA complexes in non-neoplastic tissues are carefully assessed.
In summary, SCAN1 is the first example of a human genetic disease that results from a failure to repair DNA–protein covalent complexes. SCAN1 likely arises not only from a quantitative change in Tdp1 activity but also from a qualitative change that renders the enzyme different from wild-type Tdp1 causing it to become covalently trapped on the DNA. Furthermore, since the previously described disease-causing neomorphic mutations are dominant (Antonarakis et al, 2001; Beaudet et al, 2001), the recessive nature of the neomorphic Tdp1 H493R mutation (Interthal et al, 2005a, 2005b) defines a novel mechanism for human disease.
Materials and methods
Human subjects
Patients gave informed consent approved by the Institutional Review Board of Baylor College of Medicine (H-9669, Houston, TX, USA) and the University of British Columbia (C06-0283, Vancouver, BC, Canada). The clinical data were collected from questionnaires completed by the referring primary care physician and from medical records and summaries provided by that physician.
Animal subjects
Mice used in this study were housed, bred, and killed in accordance with accepted ethical guidelines. These procedures were approved by the Institutional Review Board of Baylor College of Medicine (Houston, TX, USA, IRB protocol: AN-2983), the University of British Columbia (Vancouver, BC, Canada, Animal Care Certificate: A06-0257), and Kagoshima University (Kagoshima, Japan).
Transgenic mice
We purchased the gene trap embryonic stem (ES) cell line XD105 from BayGenomics (http://baygenomics.ucsf.edu/). XD105 was generated by integration of pGT1Lxf into intron 11 of Tdp1. This cell line was injected into albino C57 blastocysts that were then implanted into pseudopregnant mothers. The resulting chimeric mice were bred to 129/SvEv females to generate founders. The progeny from these heterozygous founders were used for all subsequent analyses. Mice were genotyped by PCR of genomic DNA using a common forward primer in intron 11 and a reverse primer in either intron 11 or in pGT1Lxf (Supplementary Table I) and data not shown. The intron 11 primer pair detects the wild-type allele but not the trapped allele, and the intron 11 forward primer with the pGT1Lxf reverse primer detects the mutant allele but not the wild-type allele. The PCR was carried out using HotMasterMix (Eppendorf) with 50 ng of genomic DNA and 10 pmol of each primer. After initial denaturation at 94°C for 2 min, amplification was performed for 40 cycles with denaturation at 94°C for 20 s, annealing at 55°C for 10 s, extension at 65°C for 90 s.
To determine if the gene trap allowed expression of full-length Tdp1 mRNA by splicing around the gene trap, we performed RT–PCR on brain RNA extracted from Tdp1−/− mice (Figure 1B). The RT–PCR primers 5′ of the trap reside in exon 2, and the primers 3′ of the trap reside in exons 12 and 16, respectively (Supplementary Table I). The RT–PCR amplification was performed as described below. To determine if the gene trap allowed expression of aberrantly spliced mRNA, we performed RT–PCR using primers residing in exons 9 and 14 (Supplementary Table I).
Bleomycin, CPT-11, etoposide, and topotecan treatment of mice
Bleomycin (Bristol-Myers Squibb, Montreal, Canada), CPT-11 (Yakult Honsha Co., Tokyo, Japan), etoposide (Novapharm, Toronto, Canada), or topotecan (LKT laboratories, Inc.) were diluted with 5% Dextrose solution prior to intraperitoneal injection. Drug or PBS was given to five wild-type and five Tdp1−/− mice according to the dosages and administration schedules indicated in the Results and Figure 5B. The mice were evaluated by electrophysiology (Supplementary data) 2 days after the last dose of CPT-11, topotecan, or bleomycin.
Comet assay
For Figure 6 and Supplementary Figure 6, the MEFs were treated with 1 μM CPT, 12.5 μg/ml bleomycin, or 12.5 μg/ml etoposide for 60 min. For Figure 6, the CPT containing medium was then replaced with fresh medium and the cells were allowed to recover for the indicated times. The MEFs were then removed from the plates with trypsin, washed with medium, resuspended in LMAgarose, and layered on agarose-coated slides cooling on ice.
Alkali comet assay (Singh et al, 1988). For CPT- or bleomycin-treated MEFs, the slides were immersed in lysis solution (2.5 M NaCl, 100 mM EDTA, 10 mM Trizma base, 1% Triton X-100, 10% dimethyl sulfoxide; pH 10) at 4°C for 1 h. After a rinse in deionized water, slides were immersed in a 4°C alkaline solution (50 mM NaOH, 1 mM EDTA, 1% dimethyl sulfoxide; pH>13) for 25 min. Electrophoresis was carried out at a constant voltage of 25 V for 25 min at 4°C. After electrophoresis, slides were neutralized in 0.4 M Tris–HCl pH 7.5.
Neutral comet assay (Olive et al, 1992). For etoposide-treated MEFs, the slides were immersed in lysis solution (50 mM EDTA, 0.5% SDS; pH 7.5) at 4°C for 1 h. After a rinse in deionized water, slides were immersed in 4°C TBE for 25 min. Electrophoresis was carried out at a constant voltage of 25 V for 25 min at 4°C.
After electrophoresis for both methods, slides were dehydrated in ice-cold 70% ethanol for 5 min and air-dried. DNA was stained with Sybr Green I. A total of 100 comets were scored for each sample using CASP (Konca et al, 2003) and statistically significant differences in the distribution of comet moments were determined using the Student t-test.
Anti-Tdp1 serum production
An anti-human Tdp1 serum was generated in rabbits against Tdp1 amino acids 1–152. An anti-mouse Tdp1 serum was generated in guinea pigs against the full-length protein (amino acids 1–608). Both antigens were produced in Escherichia coli with the pET28a expression system (Novagen). Neither antigen displayed homology to other human proteins by BLASTp. The specificity of each antiserum was confirmed by the absence of crossreactivity with recombinant Tdp1 expressed in Tdp1−/− MEFs and by competitive blocking with recombinant human or mouse Tdp1, respectively.
Immunohistochemistry and histopathology
Human and mouse brains were fixed by immersion in 10% buffered formalin or 4% PFA in PBS. The brain tissue was processed, embedded in paraffin, and cut into 8 mm sections according to standard protocols (Deguchi et al, 2003). Immunohistochemistry was carried out as previously described (Kilic et al, 2005). We used the polyclonal rabbit anti-human Tdp1 at a dilution of 1:50 and the polyclonal guinea pig anti-mouse Tdp1 at a dilution of 1:50. TUNEL detection of apoptotic cells was carried out using the ApopTag Peroxidase In Situ Apoptosis Detection Kit (Chemicon, S7100) and the tissue was counterstained with 1% methyl green.
Modified ICE assay (Subramanian et al, 1995)
Briefly, two confluent 15 cm Petridishes of telomerase immortalized SCAN1 or control human fibroblasts were treated with 20 μM CPT for 1 h. Cells were lysed in 0.8% SDS in TE and the genomic DNA was sheared with a syringe. Small aliquots of the whole cell extracts were mixed with SDS loading buffer for later analysis. The remaining cell lysates were diluted four-fold with 1% N-lauroylsarcosine and the extracts were layered onto a CsCl cushion (1.5 g/cc). After ultracentrifugation, the pellet fraction was treated with micrococcal nuclease to remove the majority of the DNA bound to the proteins. Samples were mixed with SDS sample buffer and subjected to SDS–PAGE and western blotting.
Tdp1 activity assay with mouse neurosphere cell extracts
Exponentially growing neurospheres were harvested by centrifugation, washed twice in PBS, and resuspended to 8 × 107 cells/ml in lysis buffer (10 mM Tris–HCl (pH 7.5), 50 mM KCl, 2 mM MgCl2, 1% Triton X-100, 15 mM DTT, 0.2 mg/ml PMSF, 1/1000 volume of protease inhibitor mixes Pic-D (5 mg/ml Pepstatin A, 1 mg/ml chymostatin in DMSO), and Pic-W (208 mg/ml benzamidine, 5 mg/ml aprotinin, and 1 mg/ml leupeptin in H2O)). The cells were lysed by vortexing for 1 min and the cell extracts were clarified by centrifugation.
Cell extracts were first diluted 1:2 with 2 × reaction buffer (200 mM KCl, 40 mM Tris–HCl (pH 7.5), 40 mM EDTA, 2 mM DTT) and then 10-fold serially diluted in reaction buffer. For the experiment shown in Figure 1D, 10 μl of the extract dilutions was added to 5 μl of reaction buffer containing 0.01 pmol of 32P-5′ end-labeled substrate 12-Y (a 12-mer DNA oligonucleotide with a 3′ phosphotyrosine (Interthal et al, 2005b)) and the reactions were incubated at 37°C for 30 min and stopped with an equal volume of formamide loading dye. Assays were analyzed on a 15% sequencing gel. Image retrieval and quantitation were carried out using a PhosphorImager and ImageQuant software (Amersham Biosciences).
Supplementary Material
Supplementary Information
Acknowledgments
We thank Millan Patel, Leah Elizondo, J Marietta Clewing, and Kensuke Shiga for critical reviews of this manuscript. We thank Barbara A Antalffy and Pauline Grennan for preparation of tissue; Monica J Justice, Darlene Skapura, and Wei Yu for mouse tissue; and Akiko Yoshimura and Yuko Shirahama for technical assistance. This work was supported in part by grants from the National Institute of Diabetes, Digestive, and Kidney Diseases, NIH (CFB, DLA), the New Development Award, Microscopy, and Administrative Cores of the Mental Retardation and Developmental Disabilities Research Center at Baylor College of Medicine (CFB, DLA), the National Ataxia Foundation (CFB), the Burroughs Wellcome Foundation (CFB), the Nervous and Mental Disorders and Research Committee for Ataxic Disease of the Japanese Ministry of Health, Welfare and Labor (grant 17A-1, HT, KA), Frontier Science Research Center Program of Kagoshima University (HT), and the Ministry of Education, Culture, Sports, Science, and Technology of Japan (grant 19591001, HT) and by NIH Grant GM049156 (JJC). NHLBI's Programs for Genomic Applications supported the generation of insertion trap ES cells at BayGenomics (grant U01 HL66621).
References
- Antonarakis SE, Krawczak M, Cooper DN (2001) The nature and mechanisms of human gene mutation. In The Metabolic & Molecular Bases of Inherited Disease, Scriver CR, Beaudet AL, Sly WS, Valle D, Childs B, Kinzler KW, Vogelstein B (eds), pp 343–377. New York: McGraw-Hill [Google Scholar]
- Arvidson B (1979) Distribution of intravenously injected protein tracers in peripheral ganglia of adult mice. Exp Neurol 63: 388–410 [DOI] [PubMed] [Google Scholar]
- Barthelmes HU, Habermeyer M, Christensen MO, Mielke C, Interthal H, Pouliot JJ, Boege F, Marko D (2004) TDP1 overexpression in human cells counteracts DNA damage mediated by topoisomerases I and II. J Biol Chem 279: 55618–55625 [DOI] [PubMed] [Google Scholar]
- Beaudet AL, Scriver CR, Sly WS, Valle D (2001) Genetics, biochemistry, and molecular bases of variant human phenotypes. In The Metabolic & Molecular Bases of Inherited Disease, Scriver CR, Beaudet AL, Sly WS, Valle D, Childs B, Kinzler KW, Vogelstein B (eds), pp 3–45. New York: McGraw-Hill [Google Scholar]
- Brookman KW, Lamerdin JE, Thelen MP, Hwang M, Reardon JT, Sancar A, Zhou ZQ, Walter CA, Parris CN, Thompson LH (1996) ERCC4 (XPF) encodes a human nucleotide excision repair protein with eukaryotic recombination homologs. Mol Cell Biol 16: 6553–6562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciccia A, Constantinou A, West SC (2003) Identification and characterization of the human mus81-eme1 endonuclease. J Biol Chem 278: 25172–25178 [DOI] [PubMed] [Google Scholar]
- Connelly JC, Leach DR (2004) Repair of DNA covalently linked to protein. Mol Cell 13: 307–316 [DOI] [PubMed] [Google Scholar]
- D'Arpa P, Beardmore C, Liu LF (1990) Involvement of nucleic acid synthesis in cell killing mechanisms of topoisomerase poisons. Cancer Res 50: 6919–6924 [PubMed] [Google Scholar]
- Deguchi K, Inoue K, Avila WE, Lopez-Terrada D, Antalffy BA, Quattrocchi CC, Sheldon M, Mikoshiba K, D'Arcangelo G, Armstrong DL (2003) Reelin and disabled-1 expression in developing and mature human cortical neurons. J Neuropathol Exp Neurol 62: 676–684 [DOI] [PubMed] [Google Scholar]
- Dunlop J, Corominas M, Serras F (2000) The novel gene glaikit, is expressed during neurogenesis in the Drosophila melanogaster embryo. Mech Dev 96: 133–136 [DOI] [PubMed] [Google Scholar]
- Dunlop J, Morin X, Corominas M, Serras F, Tear G (2004) Glaikit is essential for the formation of epithelial polarity and neuronal development. Curr Biol 14: 2039–2045 [DOI] [PubMed] [Google Scholar]
- El-Gizawy SA, Hedaya MA (1999) Comparative brain tissue distribution of camptothecin and topotecan in the rat. Cancer Chemother Pharmacol 43: 364–370 [DOI] [PubMed] [Google Scholar]
- El-Khamisy SF, Caldecott KW (2007) DNA single-strand break repair and spinocerebellar ataxia with axonal neuropathy-1. Neuroscience 145: 1260–1266 [DOI] [PubMed] [Google Scholar]
- El-Khamisy SF, Saifi GM, Weinfeld M, Johansson F, Helleday T, Lupski JR, Caldecott KW (2005) Defective DNA single-strand break repair in spinocerebellar ataxia with axonal neuropathy-1. Nature 434: 108–113 [DOI] [PubMed] [Google Scholar]
- Friedberg EC, Walker GC, Siede W, Wood RD, Schultz RA, Ellenberger T (2006) Disease States associated with defective biological responses to DNA damage. In DNA Repair and Mutagenesis, pp 863–1080. Washington: ASM Press [Google Scholar]
- Inamdar KV, Pouliot JJ, Zhou T, Lees-Miller SP, Rasouli-Nia A, Povirk LF (2002) Conversion of phosphoglycolate to phosphate termini on 3′ overhangs of DNA double strand breaks by the human tyrosyl-DNA phosphodiesterase hTdp1. J Biol Chem 277: 27162–27168 [DOI] [PubMed] [Google Scholar]
- Interthal H, Chen HJ, Champoux JJ (2005a) Human Tdp1 cleaves a broad spectrum of substrates, including phosphoamide linkages. J Biol Chem 280: 36518–36528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Interthal H, Chen HJ, Kehl-Fie TE, Zotzmann J, Leppard JB, Champoux JJ (2005b) SCAN1 mutant Tdp1 accumulates the enzyme–DNA intermediate and causes camptothecin hypersensitivity. EMBO J 24: 2224–2233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Interthal H, Pouliot JJ, Champoux JJ (2001) The tyrosyl-DNA phosphodiesterase Tdp1 is a member of the phospholipase D superfamily. Proc Natl Acad Sci USA 98: 12009–12014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilic SS, Donmez O, Sloan EA, Elizondo LI, Huang C, Andre JL, Bogdanovic R, Cockfield S, Cordeiro I, Deschenes G, Frund S, Kaitila I, Lama G, Lamfers P, Lucke T, Milford DV, Najera L, Rodrigo F, Saraiva JM, Schmidt B et al. (2005) Association of migraine-like headaches with Schimke immuno-osseous dysplasia. Am J Med Genet A 135: 206–210 [DOI] [PubMed] [Google Scholar]
- Konca K, Lankoff A, Banasik A, Lisowska H, Kuszewski T, Gozdz S, Koza Z, Wojcik A (2003) A cross-platform public domain PC image-analysis program for the comet assay. Mutat Res 534: 15–20 [DOI] [PubMed] [Google Scholar]
- Liu C, Pouliot JJ, Nash HA (2002) Repair of topoisomerase I covalent complexes in the absence of the tyrosyl-DNA phosphodiesterase Tdp1. Proc Natl Acad Sci USA 99: 14970–14975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu LF (1989) DNA topoisomerase poisons as antitumor drugs. Annu Rev Biochem 58: 351–375 [DOI] [PubMed] [Google Scholar]
- Matsunaga T, Mu D, Park CH, Reardon JT, Sancar A (1995) Human DNA repair excision nuclease. Analysis of the roles of the subunits involved in dual incisions by using anti-XPG and anti-ERCC1 antibodies. J Biol Chem 270: 20862–20869 [DOI] [PubMed] [Google Scholar]
- Miao ZH, Agama K, Sordet O, Povirk L, Kohn KW, Pommier Y (2006) Hereditary ataxia SCAN1 cells are defective for the repair of transcription-dependent topoisomerase I cleavage complexes. DNA Repair (Amst) 5: 1489–1494 [DOI] [PubMed] [Google Scholar]
- Nitiss KC, Malik M, He X, White SW, Nitiss JL (2006) Tyrosyl-DNA phosphodiesterase (Tdp1) participates in the repair of Top2-mediated DNA damage. Proc Natl Acad Sci USA 103: 8953–8958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nouspikel T, Hanawalt PC (2002) DNA repair in terminally differentiated cells. DNA Repair (Amst) 1: 59–75 [DOI] [PubMed] [Google Scholar]
- Ogrunc M, Sancar A (2003) Identification and characterization of human MUS81-MMS4 structure-specific endonuclease. J Biol Chem 278: 21715–21720 [DOI] [PubMed] [Google Scholar]
- Olive PL, Wlodek D, Durand RE, Banath JP (1992) Factors influencing DNA migration from individual cells subjected to gel electrophoresis. Exp Cell Res 198: 259–267 [DOI] [PubMed] [Google Scholar]
- Pommier Y (2004) Camptothecins and topoisomerase I: a foot in the door. Targeting the genome beyond topoisomerase I with camptothecins and novel anticancer drugs: importance of DNA replication, repair and cell cycle checkpoints. Curr Med Chem Anti-Canc Agents 4: 429–434 [DOI] [PubMed] [Google Scholar]
- Pouliot JJ, Robertson CA, Nash HA (2001) Pathways for repair of topoisomerase I covalent complexes in Saccharomyces cerevisiae. Genes Cells 6: 677–687 [DOI] [PubMed] [Google Scholar]
- Pourquier P, Pommier Y (2001) Topoisomerase I-mediated DNA damage. Adv Cancer Res 80: 189–216 [DOI] [PubMed] [Google Scholar]
- Sestili P, Martinelli C, Stocchi V (2006) The fast halo assay: an improved method to quantify genomic DNA strand breakage at the single-cell level. Mutat Res 607: 205–214 [DOI] [PubMed] [Google Scholar]
- Singh NP, McCoy MT, Tice RR, Schneider EL (1988) A simple technique for quantitation of low levels of DNA damage in individual cells. Exp Cell Res 175: 184–191 [DOI] [PubMed] [Google Scholar]
- Subramanian D, Kraut E, Staubus A, Young DC, Muller MT (1995) Analysis of topoisomerase I/DNA complexes in patients administered topotecan. Cancer Res 55: 2097–2103 [PubMed] [Google Scholar]
- Takashima H, Boerkoel CF, John J, Saifi GM, Salih MA, Armstrong D, Mao Y, Quiocho FA, Roa BB, Nakagawa M, Stockton DW, Lupski JR (2002) Mutation of TDP1, encoding a topoisomerase I-dependent DNA damage repair enzyme, in spinocerebellar ataxia with axonal neuropathy. Nat Genet 32: 267–272 [DOI] [PubMed] [Google Scholar]
- Vance JR, Wilson TE (2002) Yeast Tdp1 and Rad1–Rad10 function as redundant pathways for repairing Top1 replicative damage. Proc Natl Acad Sci USA 99: 13669–13674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weimer LH, Podwall D (2006) Medication-induced exacerbation of neuropathy in Charcot Marie Tooth disease. J Neurol Sci 242: 47–54 [DOI] [PubMed] [Google Scholar]
- Yang SW, Burgin AB Jr, Huizenga BN, Robertson CA, Yao KC, Nash HA (1996) A eukaryotic enzyme that can disjoin dead-end covalent complexes between DNA and type I topoisomerases. Proc Natl Acad Sci USA 93: 11534–11539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou T, Lee JW, Tatavarthi H, Lupski JR, Valerie K, Povirk LF (2005) Deficiency in 3′-phosphoglycolate processing in human cells with a hereditary mutation in tyrosyl-DNA phosphodiesterase (TDP1). Nucleic Acids Res 33: 289–297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuang Y, Fraga CH, Hubbard KE, Hagedorn N, Panetta JC, Waters CM, Stewart CF (2006) Topotecan central nervous system penetration is altered by a tyrosine kinase inhibitor. Cancer Res 66: 11305–11313 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information