Abstract
Pax6 is a lineage-restricted DNA-binding transcription factor regulating the formation of mammalian organs including brain, eye and pancreas. Pax6 plays key roles during the initial formation of lens lineage, proliferation of lens progenitor and precursor cells and their terminal differentiation. In addition to Pax6, lens fiber cell differentiation is regulated by c-Maf, Prox1 and Sox1. Crystallins are essential lens structural proteins required for light refraction and transparency. Mouse αA-crystallin represents about 17% of all crystallins at the protein level and ranks as one of the most abundant tissue-specific proteins. Lens-specific expression of this gene is regulated at the level of transcription. A promoter fragment of −88 to +46 is capable of driving lens-specific expression in transgenic mouse. Here we provide data suggesting that this lens-specific promoter fragment is comprised of multiple Pax6 and Maf-binding sites. Site-directed mutagenesis of regions within these sites resulted in partially or completely reduced promoter activities in lens cells. Co-transfections using Pax6 and c-Maf alone revealed moderate and strong activations of this promoter, respectively. In contrast to synergistic activation of αB-crystallin by Pax6 and c-Maf, Pax6 has a neutral effect on c-Maf-mediated αA-crystallin promoter activation. Chromatin immunoprecipitations established in vivo interactions of Pax6 and c-Maf with the αA-crystallin promoter in lens cells. Collectively, the present data support a molecular model in which tissue-specific expression of αA-crystallin is regulated by recruitment of Pax6 and c-Maf, two proteins regulating multiple processes of lens differentiation, to its promoter. In addition, the data suggest a molecular model of temporal and spatial regulation of αB, αA and γ-crystallin genes in mouse embryonic lens by using variants of the Pax6/ Maf regulatory module.
Keywords: lens, crystallins, differentiation, Pax6, c-Maf
Introduction
Pax6 is a lineage-restricted specific DNA-binding transcription factor regulating the formation of mammalian organs including brain, eye and pancreas.1,2 It has a particularly critical role in the development of visual systems from cells of ectodermal, neuroectodermal, and mesenchymal origin.3–5 Despite the central roles of Pax6 in these processes, only about 20 tissue-specific and tissue-restricted genes have currently been shown as directly regulated by this factor in brain, cornea, lens, retina, and pancreas,1,6 It has been proposed that Pax6 regulates expression of its direct target genes in concert with other lineage-restricted transcription factors including members of the large Maf family (MafA, MafB, c-Maf and NRL), Pax2, Prox1, Six3, Sox1 and Sox2.6–8 Expression of crystallin genes in lens provides an excellent opportunity to study the molecular mechanism of gene activation and repression by Pax6.
Lens lineage originates from a population of cells comprising the head surface ectoderm by a concerted action of transcription factors including Pax6, Otx2, and Sox2, and signaling pathways such as BMPs, FGFs, and retinoic acid.9,10 An area of the surface ectoderm in contact with the underlying optic vesicle thickens to form the lens placode and initiates expression of additional lens lineage-restricted factors including Six3, Pitx3, FoxE3, and later Prox1. In the mouse, αB-crystallin is the first and only crystallin expressed in the lens placode.11,12 As the lens placode buckles inward, it produces a recognizable depression in the exterior surface marked by expression of c-Maf, and is followed by the onset of αA-crystallin expression.11 The resulting structure, a spherical lens vesicle, is polarized. Its anterior cells retain their epithelial morphology whereas the posterior cells terminally differentiate, forming the primary lens fibers. This process is controlled by growth factors including FGF2 originating from the prospective retina, in conjunction with Prox1's critical nuclear role to induce cell cycle arrest.13 Lens fiber cell differentiation is characterized by cellular elongation and a dramatic increase in αA and αB-crystallin synthesis as well as other crystallins comprising the βγ-crystallin subfamily.6,9,10 In parallel, expression of Pax6 is reduced but not abolished in differentiating lens fiber cells and high level of c-Maf expression is maintained throughout the embryonic stages of lens differentiation and growth.14,15 Since αA-crystallin is the most abundant crystallin, representing about 17% of all crystallins16 in the newborn mouse lens, understanding the molecular basis of its temporally and spatially regulated expression in the lens is an important issue of cellular differentiation in general.
Lens-specific expression of the αA-crystallin gene is regulated at the level of transcription.6 A promoter fragment of −88 to +46 is sufficient to support lens-specific expression of the chloramphenicol acetyl transferase (CAT) reporter gene.17 Transcription factors regulating in vivo expression of this gene emerged from gene targeting studies of lens lineage-specifying genes described above. Inactivation of c-Maf,18,19 but not of Prox113 and Sox2,20 resulted in a significant reduction of both αA and αB-crystallin expression in c-Maf null lenses. Although defective fiber cell differentiation is a major phenotype in c-Maf homozygous mouse, c-Maf plays important roles in organs not expressing the αA-crystallin.21–24 Conditional inactivation of Pax6 from the lens placode completely abrogated lens development with no αA-crystallin expression.25 A c-Maf binding site (MARE) interacting with recombinant c-Maf was shown between nucleotides −110 and −98 of the extended promoter18 while a Pax6-binding site (−54/ −35) was identified earlier.26 A DNaseI footprinting study of the αA-crystallin promoter revealed binding of lens nuclear proteins to sites surrounding this Pax6-binding site and an adjacent TATA-region;27 however, interactions with lens-lineage restricted transcriptions were not further pursued. Binding of ubiquitously expressed AP-1 factor c-Jun from mouse lens epithelial cells to the downstream region (+27/ +31) was also shown earlier.28 Here, we tested the hypothesis that a lens-specific promoter fragment −88 to +46 of the mouse αA-crystallin promoter harbors novel binding sites for lens-lineage specific transcription factors since a single Pax6-binding site described above is insufficient to elicit lens-specific expression driven by the −88 to +46 promoter fragment.17,26,29 We indeed found that this region contains three novel large MAREs. There are four large Maf proteins; MafA, MafB, c-Maf, and NRL, all of which are expressed in the lens.6,9 We show that each large Maf can activate the mouse αA-crystallin promoter in transfected cells. We have previously shown transcriptional synergism between large Mafs and Pax6 proteins in the mouse αB-crystallin promoter30,31 that contrasted with transcriptional co-repression by Pax6 and Six3 of the Maf/Sox-mediated activation of the γF-crystallin promoter.31 Since αA is expressed later than αB-crystallin but prior to the onset of γ-crystallins, and αA is the most abundant of the 16 crystallins in mouse and human lens, functional interactions between Pax6 and Mafs on this gene were probed. We also present evidence for three Pax6-binding sites in the mouse αA-crystallin promoter. Using chromatin immunoprecipitations (ChIPs) we show for the first time that the αA-crystallin promoter binds both Pax6 and c-Maf in vivo. Finally, the present data led us to propose a molecular model that explains sequential activation of αB, αA and γF-crystallins in the developing mouse lens.
Results
Prediction of novel Maf and Pax6-binding sites in the mouse αA-crystallin promoter
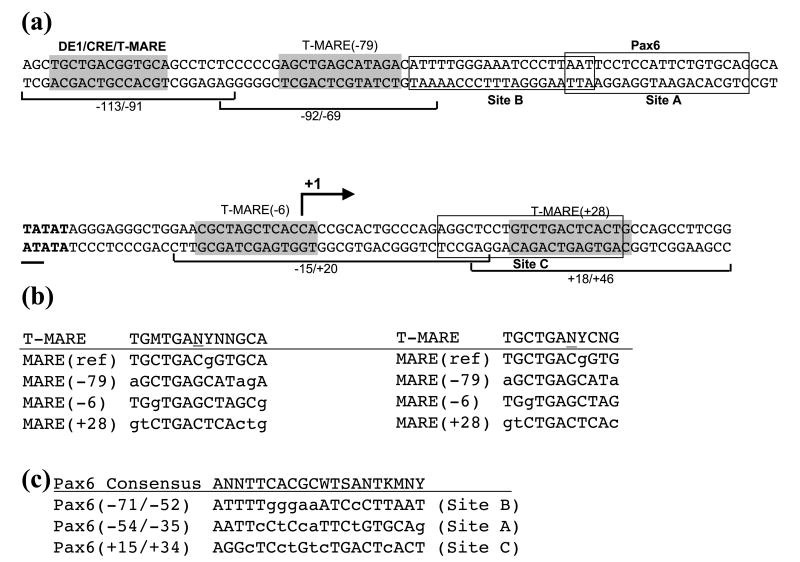
Tissue-specific gene regulation is based on a combinatorial deployment of multiple DNA-binding transcription factors with lineage-restricted expression.32 To identify putative transcription factor-binding sites residing in the lens-specific promoter region −88 to +46, we first used algorithms to predict these sites as described in Materials and Methods followed by a focused search for possible binding sites of lens lineage-specific factors including Pax6, Pitx3, Prox1, Six3, Sox1, and Sox2.6 From the first search, general homeodomain-binding sites, Ets/GATA and Smad-binding sites emerged. EMSAs were performed to assess whether proteins from these families could interact in vitro with the predicted sites and point to the possible mechanism of lens-specificity of the −88 to +46 fragment shown in transgenic mice;17,29 however, no positive data were obtained (data not shown). Next, using a consensus binding sequence 5′-TGMTGANYNNGCA-3′ for T-MARE33 and using a consensus sequence 5′-TGCTGANYCNG-3′ obtained from an alignment of 13 well-established large Maf binding sites34 we found three novel putative Maf-binding sites as shown in Figure 1. These sites are called MARE (−79), MARE(−6) and MARE(+28). Since c-Maf serves as an important factor regulating expression of mouse αA-crystallin in vivo,18,35 but its only known binding site (−110 to −98, DE1/CRE/MARE)18 is not essential for establishing lens-specific expression,17,29 prediction of additional Maf-binding sites in the −88 to +46 promoter fragment was considered significant for its transcriptional regulation. In addition, this region may contain two novel Pax6-binding sites, −71 to −51 (site B) and +15 to +34 (site C), as indicated by the sequence conservation between these sequences and the optimal binding sites for the Pax6 paired domain, P6CON, 5′-ANNTTCACGCWTSANTKMNY-3′ (Figure 1).36
Figure 1.
Diagrammatic representation of large Maf and Pax6-binding sites in the mouse αA-crystallin promoter. (a) The mouse promoter region is shown from positions −111 to +46. The previously established DE1/CRE/T-MARE and Pax6-binding site A are shown in bold. Novel T-MARE(−79), T-MARE(−6) and T-MARE(+28) were numbered according to the nucleotide (underlined) in the center of the 13 bp consensus binding, TGMTGANYNNGCA. A similar alignment with the consensus binding site for Pax6, ANNTTCACGCWTSANTKMNY, predicts two novel binding sites (B and C, boxed). K=G/T; M=A/C; N=A/C/G/T; S=C/G; W=A/T and Y=C/T. (b) Alignments of each αA-crystallin T-MARE with a 13 (left) and an 11 -base-pair (right) consensus MAREs. (c) Alignments of Pax6-binding sites A, B and C with their consensus binding sites. Mismatched nucleotides are shown in lower case letters.
Large Mafs activate the mouse αA-crystallin promoter
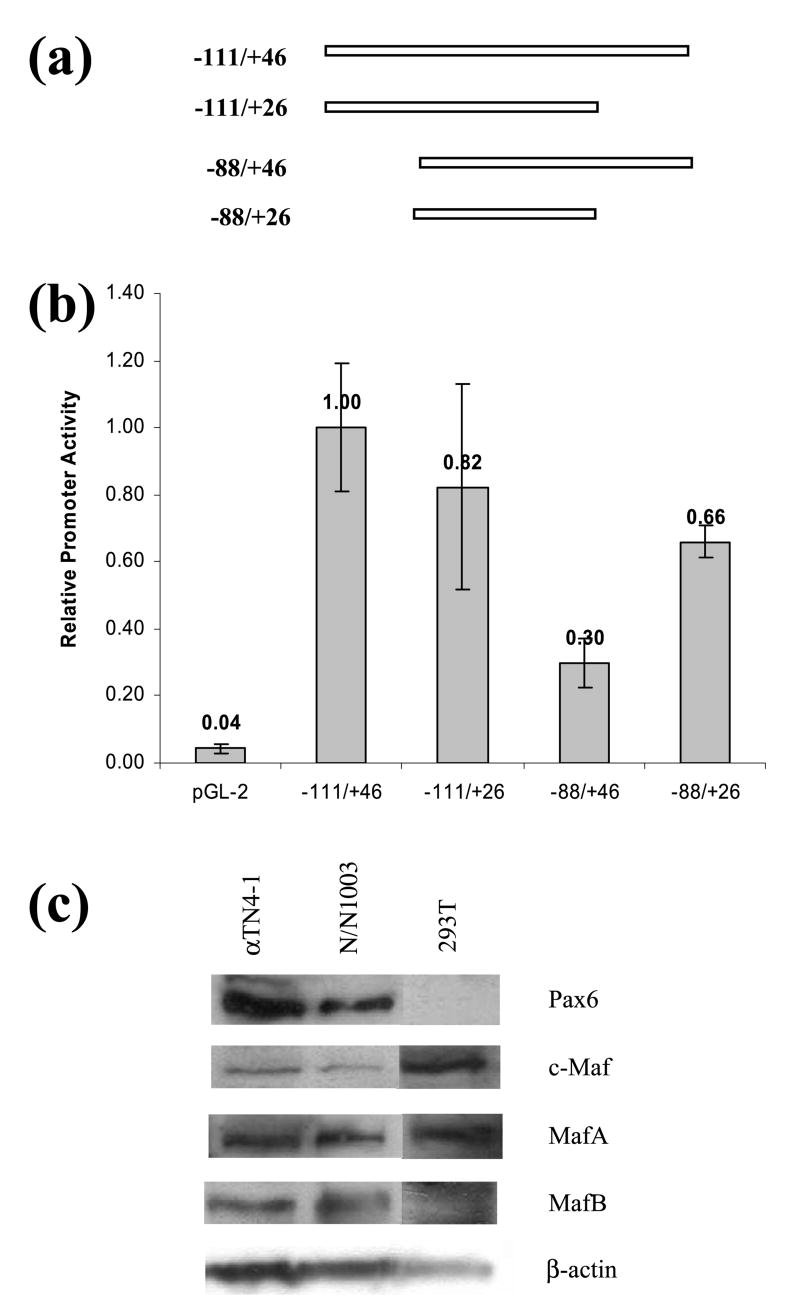
To initially test our hypothesis, we assessed the activity of the −111 to +46 promoter region and loss of the 5′ and 3′-MAREs, i.e. DE1/CRE/MARE and MARE(+28). Four fragments, −111 to +46, −88 to +46, −111 to +24, and −88 to +24, that drive the expression of luciferase in cultured lens cells were tested as shown in Figure 2(a). The results (Figure 2(b)) revealed reduced activities of truncated promoters. Even the shortest fragment, −88 to +26, with two predicted MAREs and Pax6-binding sites had still appreciable promoter activity. Western immunoblotting was performed to evaluate expression of Pax6, c-Maf, MafA and MafB in lens cultured cells, mouse αTN4-1 and rabbit N/N1003A, and in 293Tcells used for transfections. Expression of these proteins was detected in all instances except for no expression of Pax6 in 293T cells. Thus, both cultured lens cell epithelial lines, αTN4-1 and N/N1003A, express all important lens lineage specific transcription factors Pax6, MafA, MafB and c-Maf.
Figure 2.
Promoter activity of the mouse αA-crystallin fragment −111/ +46 and its truncations in lens cells. (a) A diagrammatic representation of the wild-type and truncated promoters cloned in the pGL2 vector with the firefly luciferase gene. (b) Relative promoter activities of −111/ +46, −111/ +26, −88/ +46, and −88/ +26 promoter fragments in cultured lens cells normalized against the activity of the −111/ +46 promoter. The transfections were performed in hexaplets and normalized using Renilla luciferase as an internal control. The results are shown as means with standard deviations indicated by error bars. (c) Results of Western immunobloting of Pax6, c-Maf, MafA and MafB in lens cultured (αTN4-1 and N/N1003A) and 293T cells. An immunoblot with β-actin is shown as a loading control.
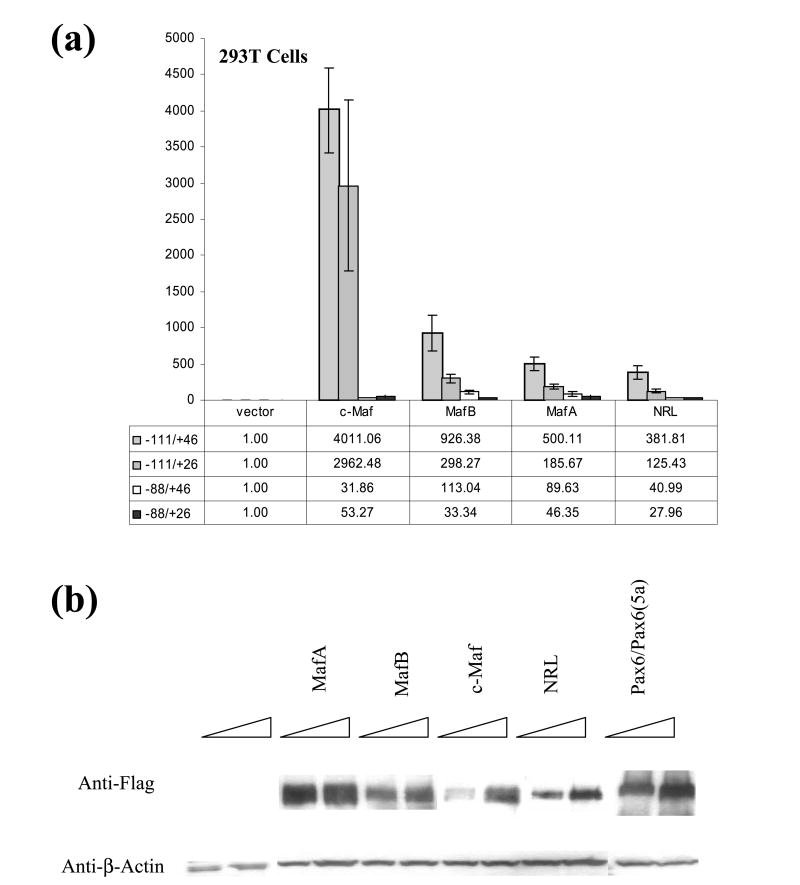
To determine whether the wild-type −111 to +46 promoter and its truncated fragments could be activated by large Mafs, we performed a series of transient cotransfections in 293T cells using individual expression plasmids encoding MafA, MafB, c-Maf, and NRL. The wild-type promoter was strongly activated by each Maf in the order of c-Maf > MafB > MafA > NRL (see Figure 3). Deletion of DE1/CRE/MARE decreased large Maf's co-activations to 18, 3, 3 and 11% of normal activity of MafA, MafB, c-Maf and NRL, respectively. In contrast, deletion of a region containing downstream MARE(+28) had the least dramatic impact with 37, 32, 74, and 33% of normal activities of co-tranfected MafA, MafB, c-Maf and NRL, respectively. The promoter fragment lacking both DE1/CRE/MARE and MARE(+28), −88 to +26, was also activated by each large Maf (see Figure 3(a)). Expression of ectopic Mafs in transfected 293T cells was assessed by Western blots and M2 anti-flag antibody (Figure 3(b)). From these data, we concluded that the mouse αA-crystallin promoter fragment, −111 to +46, possesses multiple MAREs and that c-Maf was the strongest activator of αA-crystallin promoter activity in these transfection assays.
Figure 3.
Transcriptional activation of four mouse αA-crystallin promoter fragments by individual c-Maf, MafB, MafA and NRL. (a) Activities of a series of αA-crystallin promoter fragments in the presence of 200 ng of each individual cDNA encoding c-Maf, MafB, MafA and NRL in transiently co-transfected 293T cells. The experiments were performed and evaluated as described in the legend to Figure 2(b). The “basal” normalized activity of −111/ +46, −111/ +26, −88/ +46 and −88/ +26 promoter fragments compared to the wild-type fragment −111/ +46 in 293T cells were 1.0, 1.43, 1.5 and 0.76, respectively. Activation potentials of individual Mafs are expressed as ratios in the presence of Maf divided by the activity in the presence of identical amounts of empty vector, pKW10. (b) Western immunoblotting to demonstrate expression of ectopic MafA, MafB, c-Maf and NRL proteins in 293T cells. 293T cells were transfected with 80 ng of expression plasmid and 20 and 40 μg of whole cell extracts were loaded in adjacent lanes. Expression of Pax6/ Pax6 (5a) is also shown as this experiment relates to data shown in Figure 9. Expression of β-actin was used as a loading control.
Identification of Maf and Pax6-binding sites in vitro in the mouse αA-crystallin promoter
EMSAs and DNase I footprinting were used to examine in vitro protein–DNA interactions of the mouse αA-crystallin promoter. The lens nuclear proteins were incubated with a series of four probes −113/ −91, −92/ −69, −15/ +20, and +18/ +46 harboring putative MAREs (see Figure 1) followed by gel electrophoresis. Each probe formed a broad band of overlapping specific complexes (Figure 4, lanes 2, 7, 13 and 19) that were abolished or reduced in the presence of oligonucleotides containing consensus T-MARE (lanes 4, 10, 16 and 22) but less in the presence of mutated T-MARE sequences (lanes 5, 11, 17 and 23). The data suggested that each probe could form specific complexes corresponding to activities of Maf proteins as shown directly by a similar probe −113/ −91 elsewhere.18 Results of specific cross-competitions of this sequence with probes −92/ −69, −15/ +20 and +18/ +46 are shown in Figure 4, lanes 9, 15 and 21, respectively.
Figure 4.
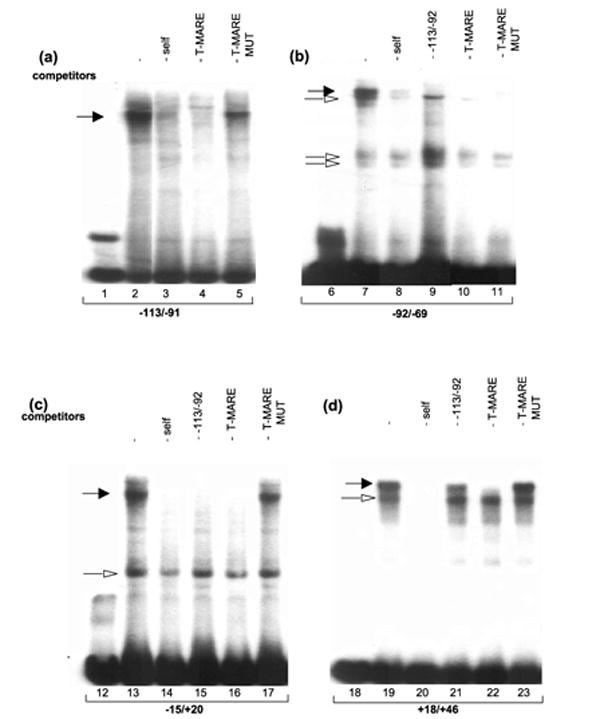
In vitro DNA-protein interactions. EMSAs with four probes derived from the mouse αA-crystallin promoter (see Figure 1) incubated with lens nuclear proteins. (a) Interaction of lens nuclear proteins (αNE) with oligonucleotide probe −113/ −91, (b) probe −92/ −69 (lanes 6 to 11), (c) probe −15/ +20 (lanes 12 to 17) and (d) probe +18/ +46 (lanes 18 to 23). Specific oligonucleotide competitors were included in lanes 3–5, 8–11, 14–17, and 20–23 as indicated. Specific complexes containing Maf-like activities and other specific complexes are labeled by filled and open arrows, respectively.
To identify the regions of the mouse αA-crystallin promoter interacting with Pax6 proteins, we used DNaseI footprinting and radioactively end-labeled promoter fragments −166 to +46. We assessed the interaction of both variants of Pax6: Pax6 containing the 128 amino acid residue paired domain (PD), and Pax6 (5a) containing the alternatively spliced 142 amino acid residue extended paired domain (PD5a). The comparison of protected areas on both lower (Figure 5(a)) and upper strand (Figure 5(b)) using proteins containing only Pax6 PD (lanes 4 and 5) or both the PD and the homeodomain (HD) confirmed that the HD restricts DNA-binding properties of both the PD and the PD(5a), in agreement with earlier studies.6 Strong binding of Pax6 proteins with both the PD and the HD (Figure 5(a), lanes 11 and 12, and Figure 5(b), lanes 11 and 12) overlapped the positions depicted in Figure 1. From these data we concluded that the αA-crystallin promoter possesses three Pax6-binding sites, labeled Pax6-site A, B and C. In addition, binding of Pax6 was found in the region −138 to −110, labeled D (see Figure 5(a) and (b), lanes 11 and 12).
Figure 5.
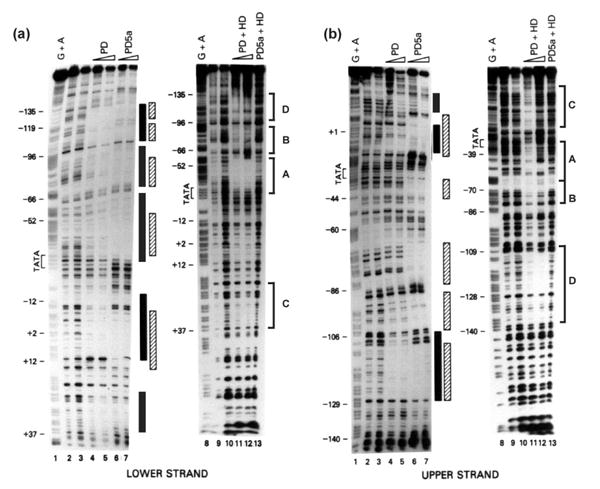
Interactions of recombinant Pax6 proteins with the mouse αA-crystallin promoter fragment using DNase I footprinting. Recombinant Pax6 PD, PD5a, PD+HD, and PD5a+HD proteins were incubated with −166 to +46 end-labeled probes and digested in the presence of DNase I. (a) Binding to the lower strand. (b) Binding to the upper strand. Areas of interaction for Pax6 PD (filled boxes), and Pax6 PD5a (shaded boxes). Four binding sites of Pax6 PD+HD proteins (brackets) were labeled A, B, C, and D (see Figure 1 for additional information).
To assess if Pax6 proteins present in lens nuclear extracts can bind to sites A, B and C, we performed a series of EMSAs. The Pax6-site A oligonucleotide (−59/ −29) was incubated with rabbit and mouse lens nuclear extracts. Three major specific and several minor complexes were detected (Figure 6(a), lanes 2 to 4). Two of them were reduced in the presence of Pax6-specific antibody recognizing its DNA-binding PD (compare lanes 4 and 5). Next, we examined if an addition of an excess of an oligonucleotide containing “optimal” Pax6-binding site, P6CON, could prevent formation of these complexes. Indeed, two complexes inhibited by the anti-Pax6 antibody were also significantly abolished in the presence of P6CON (Figure 6(b), lane 8) but not in the presence of oligonucleotides with PU.1 (lane 9) and GATA3 (lane 10) binding sequences. We next incubated an oligonucleotide −88 to −56 containing Pax6-binding site B with mouse lens nuclear extracts and identified two specific complexes (Figure 6(c), lane 2), which were abolished in the presence of P6CON but not in the presence of oligonucletides with GATA3 and NF-κB binding sites (Figure 6(c), compare lanes 2 and 7). Note that the faster migrating Pax6-containing complex was more efficiently inhibited compared to the slower Pax6-complex, likely containing additional protein(s). Finally, we incubated an oligonucleotide probe +18/ +46 with lens nuclear proteins; however this probe formed a number of slow and closely migrating complexes that were not competed in the presence of P6CON (data not shown). It also bound recombinant Pax6 proteins in EMSAs (data not shown). Earlier, binding of lens nuclear proteins to a region of +24 to +43 was shown by DNase I footprinting.27 Thus, binding of Pax6 to site C was not conclusively shown. Together, these data show that the mouse αA-crystallin promoter contains at least two Pax6-binding sites A and B.
Figure 6.
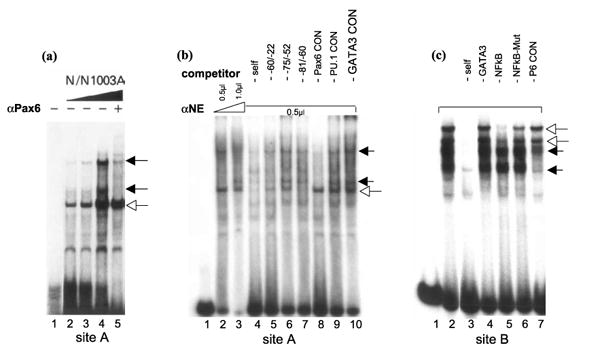
EMSAs with two probes derived from the mouse αA-crystallin promoter (see Figure 1) incubated with lens nuclear proteins. (a) An oligonucleotide −59/ −29 (site A) was incubated with increasing amounts of rabbit lens nuclear extracts from N/N1003A cells (lanes 2 to 4) and in the presence of a specific anti-Pax6 antibody recognizing Pax6 PD (lane5). (b) An oligonuclotide −59/ −29 (site A) was incubated with increasing amounts of mouse lens nuclear extracts from αTN4-1 cells (lanes 2 and 3) and in the presence of approximately 50 molar excess of indicated specific oligonucleotide competitors (lanes 4 to 10). (c) An oligonucleotide −88/ −56 (site B) was incubated with mouse lens nuclear extracts from αTN4-1 cells (lanes 2) and in the presence of approximately 50 molar excess of indicated specific oligonucleotide competitors (lanes 3 to 7). Specific complexes containing Pax6 proteins and other specific complexes are labeled by filled and open arrows, respectively.
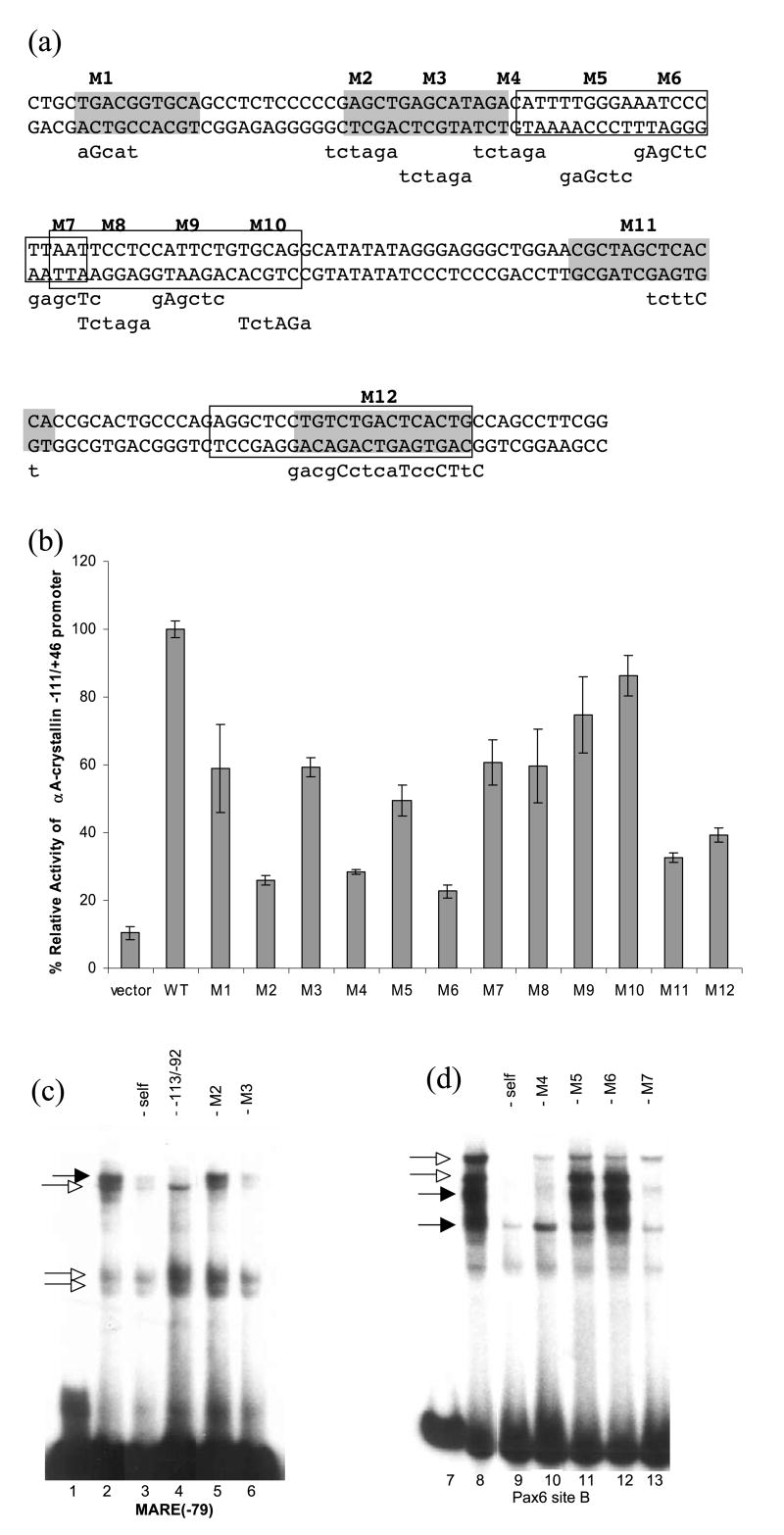
Site-directed mutagenesis of the mouse αA-crystallin promoter fragment −111 to +46
To demonstrate the functional roles of sequences including MAREs and Pax6-binding sites of the αA-crystallin promoter, we performed site-directed mutagenesis of the promoter and tested corresponding promoter activities in cultured lens cells. Twelve mutants are shown in Figure 7(a) and the results of transfections are given in Figure 7(b). Promoter activities were reduced but not abolished for all 12 tested mutations, which overlapped with the binding sites of large Maf and Pax6 proteins (see Figure 7(a)). Note that mutation M12 affected both MARE and Pax6-binding sequences. To test whether these mutations indeed influenced binding of c-Maf and Pax6, we tested mutations M2 and M3, in MARE(−79) and M4 to M7, in Pax6-binding site B. Mutations M2 and M6 most strongly reduced promoter activity. EMSAs showed that M2 could not reduce formation of Maf-like complexes (Figure 7(c), compare lanes 2 and 5). Similarly, M6 could not inhibit formation of Pax6-containing complexes using Pax6-binding site B (Figure 7(d), compare lanes 8 and 12). In addition, mutation M3 (lane 6) retained some ability to compete Maf-like complex as it caused only a 40% reduction of the αA-crystallin promoter activity (Figure 7(b)).
Figure 7.
Activities of a series of mouse αA-crystallin promoter mutants in cultured lens cells. (a) Site-directed mutagenesis of the promoter fragment −111 to +46. Twelve block mutations are labeled M1 to M12. Four T-MARE (shaded boxes) and three Pax6 (open boxes) binding sites are labeled. Nucleotide sequences used for mutagenesis are shown under the promoter sequence. (b) Relative promoter activities of M1 to M12 compared to the −111 to +46 promoter (WT). The results were calculated as described in the legend to Figure 2. (c) EMSAs using MARE(−79) (see Figures 1(a) and 4(b)) and mouse lens nuclear extract (lane 2) in the presence of about 50-fold excess of indicated oligonucleotide competitors harboring mutations M2 and M3 shown in (a) (lanes 5 and 6). (d) EMSAs using Pax6 site B (see Figures 1(a) and 6(c)) and mouse lens nuclear extract (lane 8) and in the presence of about 50-fold excess of indicated oligonucleotide competitors harboring mutations M4, M5, M6 and M7 shown in (a) (lanes 10 to 13).
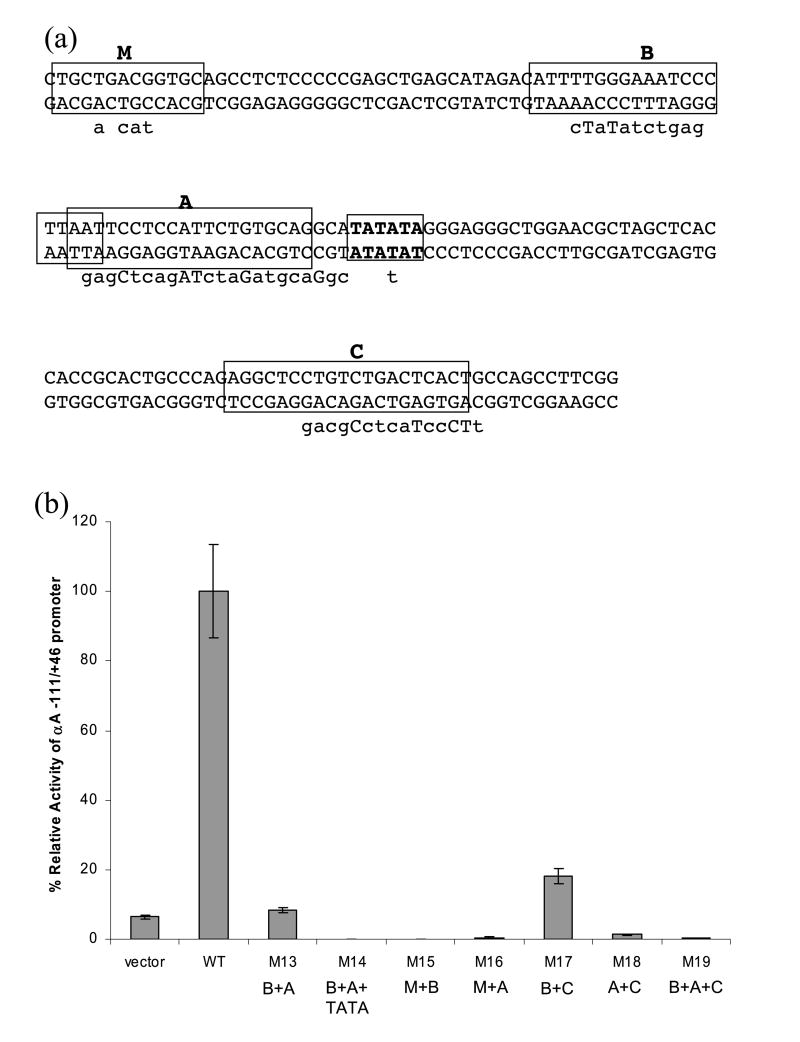
We then hypothesized that multiple mutations would result in the additional loss of promoter activities. To test this, we generated five double mutants and two triple mutants (see Figure 8(a)) and determined the promoter activities (see Figure 8(b)) as described above. We focused on those combinations including DE1/CRE/MARE and Pax6-binding site B as we could correlate the impact of these mutations with their previous studies in transgenic mice.29 Two double mutations (M15 and M16) and both triple mutations (M14 and M19) eliminated the promoter activity. M15 corresponded to the double mutation resulting in loss of promoter activity tested in transgenic mice.29 These double mutations had impaired MARE and either Pax6-site B or A. Two double mutations, M13 (affecting Pax6 sites A and B) and M17 (affecting Pax6 sites B and C), resulted in 8 and 18% of activity of the wild-type promoter, respectively. A mutation, M19, affected both Pax6-binding sites A and B established in Figures 5 and 6, and a site C, established only by use of recombinant Pax6 proteins (see Figure 5).
Figure 8.
Activities of a series of mouse αA-crystallin promoter double and triple mutants in cultured lens cells. (a) Multiple site-directed mutagenesis of the promoter fragment −111 to +46. Five double and two triple block mutations are labeled M14 to M20. DE1/CRE/T-MARE (M), two Pax6 (sites A and B), and overlapping Pax6/MARE (site C) binding sites are boxed. Nucleotide sequences used for mutagenesis are shown under the promoter sequence. (b) Relative promoter activities of M14 to M20 compared to the −111 to +46 promoter (WT). The results were calculated as described in the legend to Figure 2.
Activation of the mouse αA-crystallin promoter by transcription factors co-expressed in developing lens epithelium and in lens fibers
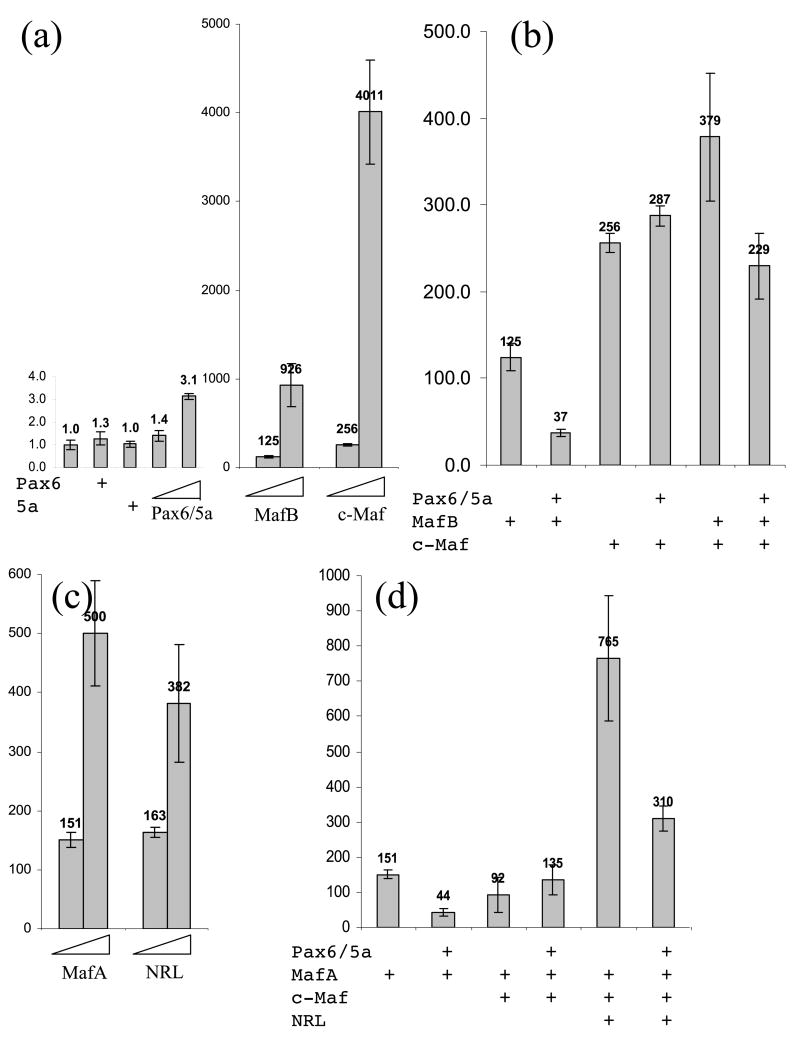
We have recently shown that mouse αB-crystallin, expressed earlier than αA-crystallin, is synergistically activated by Pax6 and large Maf proteins.30,31 In contrast, the γF-crystallin gene, which is activated by a combination of Maf and Sox1/2 proteins, after αA-crystallin, was repressed by Pax6.31,37 Here we cotransfected the wild-type αA-crystallin promoter with Pax6, Pax6 (5a), Maf B, and c-Maf as the onset of αA-crystallin expression in lens pit could be triggered by Pax6 and emerging MafB and c-Maf. Pax6 proteins alone weakly activated the mouse αA-crystallin promoter in 293T cells (see Figure 9(a)). A similar result was reported earlier in COP-8 and C33-A cells.6,26 In contrast, MafB and c-Maf strongly activated this promoter in a concentration-dependent manner (Figure 9(a)). When combinations of these factors were tested, the results showed moderate repression of MafB and MafB/c-Maf-stimulated promoter activities and a marginal increase of the promoter activity by Pax6 proteins in combination with c-Maf (see Figure 9(b)). Both MafA and NRL also activated the promoter in a dose-dependent manner (see Figure 9(c)). Similar results, i.e. weak repression of MafA and MafA/c-Maf/NRL-mediated transcriptional activation of the −111 to +46 promoter by Pax6 proteins and insignificant gain of MafA/ c-Maf-stimulated promoter activity by Pax6 proteins were found (Figure 9(d)). Next, we tested these combinations in cultured lens epithelial cells. Neither factor alone or in combination was able to activate this promoter (see Figure 1, Supplementary Data) likely due to the high basal activity of this promoter in lens cells compared to 293T cells due to the expression of endogenous Pax6, MafA, Maf B and c-Maf proteins (see Figure 2(c)).
Figure 9.
The regulation of mouse αA-crystallin promoter activity by transcription factors expressed in lens epithelium and in lens fibers. The promoter fragment −111 to +46 was co-transfected with cDNAs encoding Pax6/Pax6(5a), MafB, c-Maf, MafA, and NRL as indicated in 293T cells. (a) Results showing concentration-dependent activation by Pax6/Pax6(5a) (100/12.5 and 200/25 ng), MafB (80 and 200 ng) and c-Maf (80 and 200 ng). (b) Results of co-transfections of transcription factors, Pax6, MafB and c-Maf, co-expressed in lens epithelium. (c) Results showing concentration-dependent activation by MafA (80 and 200 ng) and NRL (80 and 200 ng). (d) Results of co-transfections of transcription factors, Pax6, MafA, c-Maf, and NRL, co-expressed in lens fiber cells.
Chromatin immunoprecipitations of transcription factors interacting with the mouse αA-crystallin promoter in vivo
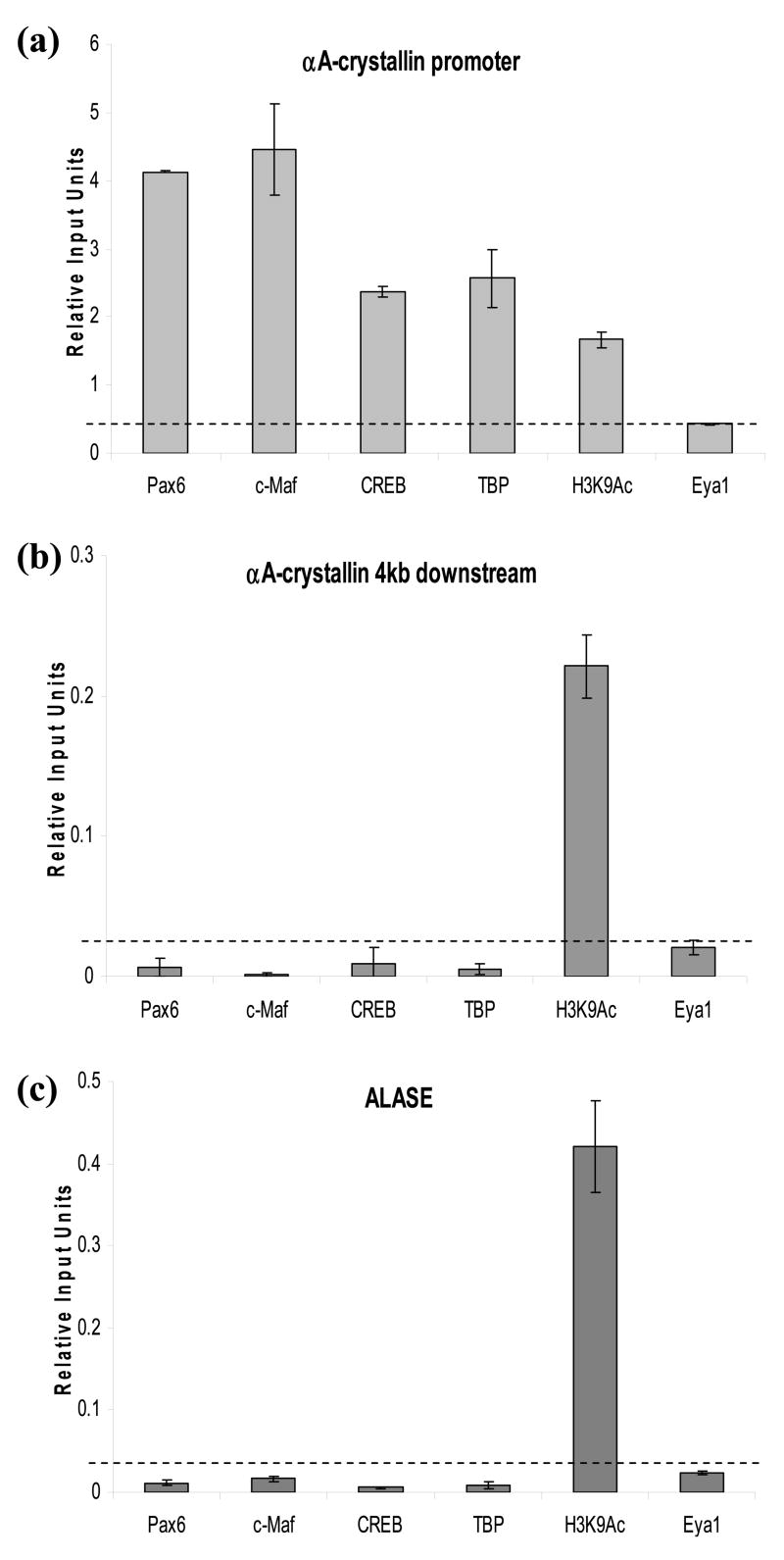
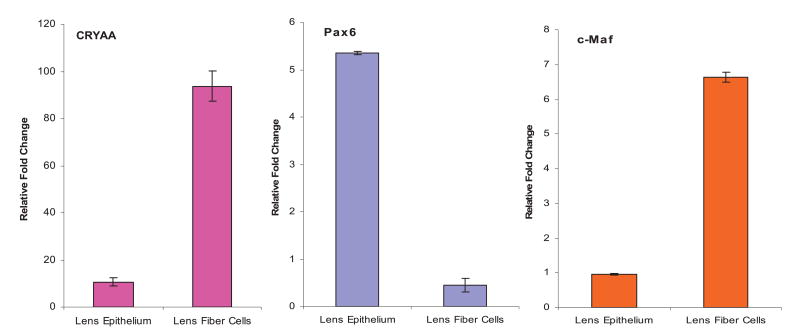
Interactions of Pax6, c-Maf, and CREB with the endogenous αA-crystallin promoter in lens cells were determined using quantitative ChIPs.38–40 Binding of general transcription factor TFIID/TBP to the promoter and transcription factor Eya1 expressed in lens was assessed in parallel. We also tested modification of histones using the anti-H3 K9 antibody, which recognizes acetylated lysine K9 of histone3. Our results (see Figure 10(a)) revealed enrichment of Pax6, c-Maf, CREB, and TFIID/TBP but not of Eya1 in the αA-crystallin promoter region in vivo. In contrast, these factors did not exhibit enrichments in the region located approximately 4 kb downstream from the promoter (Figure 10(b)) or on erythroid-specific gene ALASE (Figure 10(c)). Interestingly, no enrichments of MafA, MafB, or NRL were detected in these tests (data not shown). Based on these findings, we evaluated levels of expression of all large Mafs, MafA, MafB, c-Maf and NRL, by qRT-PCR as they are expressed in the lens with distinct temporal and spatial features.14,18,19,41–43 In one-day old mouse lens, we found significant expression of c-Maf, but not of other Mafs (data not shown). In contrast, expression of both MafA and NRL was detected in the retina and expression of MafB was found in the mouse cornea (data not shown). Next, we determined expression of these genes in the microdissected two-day old rat lens epithelium and fibers (Figure 11). As expected,14,15 the lens epithelium expressed higher levels of Pax6 compared to lens fibers (Figure 11), and, conversely, higher levels of c-Maf were found in lens fiber cells compared to lens epithelium (Figure 11). Expression of αA-crystallin between lens epithelium and fibers is shown in parallel. Thus, these data show in vivo association of the αA-crystallin promoter with Pax6, c-Maf, CREB, and TFIID/TBP and they demonstrate significant levels of expression of both Pax6 and c-Maf in one-day old mouse lens but not of any other large Mafs.
Figure 10.
Pax6, c-Maf, CREB and TBP proteins interact with the mouse αA-crystallin promoter in vivo. Quantitative ChIPs using antibodies recognizing Pax6, c-Maf, CREB, TBP, Eya1, and H3K9Ac. (a) Primers amplified the αA-crystallin promoter region and (b) the 4 kb downstream sequence of the promoter in formaldehyde crosslinked αTN4-1 lens cells. (c) Interactions with the promoter region of 5-aminolevulinate synthase (ALASE), are shown as “irrelevant” gene control. Values shown are normalized on a standard curve.38–40 Relative input units were calculated as described in Materials and Methods. Dotted lines represent averaged background signal in the presence of normal rabbit IgG for each primer set/site. Average “enrichments” are shown including standard deviations. Each experiment was performed using two independent chromatin preparations.
Figure 11.
Quantitative RT-PCR analysis of αA-crystallin, Pax6 and c-Maf expression in microdissected two-day old rat lens epithelia and lens fibers. Expression levels were normalized using standard curves for each amplicon and are expressed relative to the CCNI gene.
Discussion
The present study had two goals. First, to identify binding sites for lens lineage-restricted transcription factors in the lens-specific promoter fragment −88 to +46 of the mouse αA-crystallin. Second, to investigate the molecular mechanism by which activation of this gene occurs by focusing on in vivo protein-DNA interactions and establishing a testable model for sequential activation of αB, αA and γ-crystallin expression in the developing embryonic lens.
Previous studies have suggested that Pax6 regulates tissue-specific gene expression via specific DNA-mediated interactions with other lineagerestricted transcription factors. In lens, cooperative interactions between Pax6 and Sox2 were shown to trigger expression of chicken δ1-crystallin in the lens precursor cells forming the lens placode.7,44 In addition, Pax6 in combination with MafB and retinoic acid receptors RARβ/RXRβ or Pax6 with c-Maf synergistically activated the mouse αB-crystallin promoter.30,31 Similar studies of pancreas-specific expression of insulin, glucagon, and somatostatin revealed positive effects of Pax6 on each gene45 and cooperativity between MafA and Pax6 on the glucagon promoter.46 Moreover, the highest activation potential of MafA was found among other insulin regulatory factors.47 It is important to note that other tissue-restricted genes including BETA-2/NeuroD, Cdx2/3, Nkx2.2 and Pdx-1 are involved in the regulation of these genes in pancreas.48 In corneal epithelium, Pax6 functionally interacts with AP-2α to regulate expression of gelatinase B/MMP9.49 The present data show that the mouse αA-crystallin promoter is comprised of an array of multiple Pax6 and Maf-binding sites. The finding of both Pax6 and Maf-binding sites in virtually all crystallin promoters and/or lens-specific enhancers indicates common mechanisms to achieve both high level and lens-specific expression. Moreover, studies of invertebrate crystallin promoters suggest similar mechanisms evolutionarily conserved although vertebrate and invertebrate crystallins are structurally unrelated.50,51
The present data suggest that the mouse αA-crystallin promoter fragment −111 to +46 contains four MAREs and at least two Pax6-binding sites. Earlier studies have identified a distal DE1/CRE/MARE and an internal Pax6-binding site A.18,26 Present data show that the distal region actually contains a tandem of MAREs followed by a tandem of Pax6-binding sites (see Figure 12). Mutagenesis of each novel MARE and Pax6-binding site resulted in the loss of promoter activity in transfected lens cells. Disruption of the distal promoter region harboring a tandem of MAREs was achieved by inserting an additional Pax6 consensus-binding site between two MAREs. This insertion dramatically changed the expression pattern of the −366/ +46 promoter fragment in transgenic mice. The expansion of expression from lens fibers into lens epithelium52 provided in vivo evidence that the integrity of distal MAREs is critical for promoter function. Transgenic constructs bearing mutations in either DE1/CRE/T-MARE or Pax6-binding site B of the −111/ +46 promoter fragment only marginally influenced lens expression of the CAT reporter gene in adult lenses.29 In contrast, simultaneous inactivation of both sites abolished expression of CAT in transgenic lenses.29 Our results are consistent with these data, demonstrating reduction of promoter activity when individual sites were mutated and a complete loss of activity of the double mutation M15. In addition, similar mutations (M16) affecting a different pair of Pax6 and Maf-binding sites also resulted in the abolition of promoter activity. It remains to be tested whether other combinations of double Maf and Pax6-binding site mutations are also effective in abolishing the αA-crystallin promoter activities.
Figure 12.
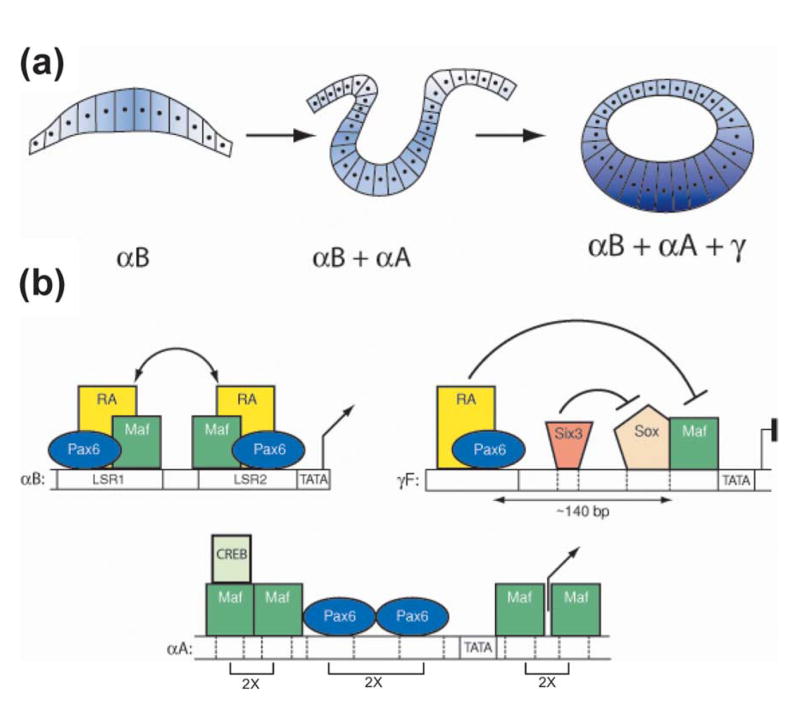
Molecular model of developmentally controlled expression of αB, αA, and γ-crystallins in mouse embryonic lens. (a) At mouse E9.5, lens placode (left panel) is formed from the surface ectoderm and expresses the αB-crystallin. At E10.5, in the invaginating lens placode (middle panel), αA-crystallin expression is triggered. From E11.5 to 13.5, primary lens fiber cells are formed from the posterior part of lens vesicle (right panel). The process is accompanied by dramatic upregulation of αA, β, and γ-crystallins in these cells. (b) Specific organization of Maf and Pax6-binding sites in crystallin promoters may regulate their temporal pattern of expression. Regulation of the mouse αB-crystallin gene (left panel). Synergistic interactions (double arrows) between Pax6, retinoic acid activated receptors (RA), and early expressed Maf proteins (c-Maf and MafB) interacting with over-lapping sites in lens specific regions LSR1 and LSR2. Regulation of the mouse αA-crystallin gene (middle panel). Moderate expression of this gene in lens pit and vesicle requires both Pax6 and Mafs. Pax6 might play an epigenetic role in opening the promoter chromatin structure. As expression of c-Maf increases in the differentiating primary lens fibers, two tandems of Maf-binding sites separated by a Pax6 tandem become fully occupied, resulting in dramatic upregulation of this gene from E11.5. Binding of CREB to the promoter agrees with earlier studies26 and ChIP data shown in Figure 10. Recruitment of CBP by c-Maf to this promoter has been shown earlier.64 Regulation of the mouse γ-crystallin genes (right panel). Expression of γ-crystallins is repressed by simultaneous action of Pax6 and Six3 protein bound distally (about 140 bp between Pax6-binding site and MARE) from the Maf binding site until the onset of lens fiber cell differentiation. Repression is relieved as a result of reduced amounts of both Pax6 and Six3 proteins, and upregulation of c-Maf in primary lens fibers.
Site-directed mutagenesis of each Pax6-binding site resulted in a reduction of αA-crystallin promoter activities, indicating that Pax6-binding is critical for its activity in lens epithelial cells. To demonstrate direct interactions of Pax6 and c-Maf with the endogenous αA-crystallin loci in cultured lens cells, we performed qChIPs. The results showed that the αA-crystallin promoter was occupied by these factors in vivo and was also enriched for CREB and TFIID/TBP. Acetylation of lysine 9 on histone H3 indicates open chromatin in the promoter. We propose that expression of Pax6 prior to c-Maf serves dual functions: Pax6 directly regulates c-Maf expression in the lens,53 and binding of Pax6 to the αA-crystallin promoter might function to open its chromatin structure.
Here we show that c-Maf is a much stronger transcriptional activator of the mouse αA-crystallin promoter compared to MafA and MafB, and its activation is not inhibited by Pax6. While c-Maf activated this promoter 256-fold (Figure 9), the mouse αB-crystallin promoter was activated only tenfold in a similar assay.31 Analysis of c-Maf null mouse lenses at E11.5 revealed 99% reduction of the αA-crystallin transcripts, directly showing that MafA and MafB do not compensate for the loss of c-Maf.18 No significant expression of MafA, MafB, and NRL by qRT-PCR was shown in one-day old mouse lens. These results were confirmed by Western immunoblot analysis of mouse lens nuclear proteins detecting c-Maf and Pax6 but not MafA and MafB (data not shown). Taken together these results show that c-Maf is the key activator of the mouse αA-crystallin due to its specificity,18 high-level of expression (Figure 11) and ability to most strongly activate its promoter (Figures 3 and 9).
Previous studies of the chicken αA-crystallin gene suggested that MafA/L-Maf is the key activator of this gene, acting downstream of Pax6.35,54,55 Genetic analysis in mouse provides evidence that loss of c-Maf, resulting in the dramatic loss of αA-crystallin expression in c-Maf null lenses, cannot be compensated by other Maf genes. There could be additional differences between the moderately conserved orthologous mouse and chicken αA-crystallin promoters56 and to date only a single T-MARE (−108/ −97) has been shown in the chicken αA-crystallin promoter fragment −162 to +44.54,55 Additional studies of the regulation of chicken αA-crystallin will be required to clarify these issues.
The spatiotemporal expression patterns of 16 crystallin genes can originate from specific placements of cis-regulatory sites interacting with a sparse number of lens lineage-specific transcription factors.6,7,57 Three crystallin promoters, αA, αB, and γF, differently responded to Pax6 and Mafs in transfection assays and these data correlate with the onset of their expression in the embryonic mouse lens (see Figure 12). Mouse αB-crystallin is first expressed in the lens placode due to synergistic interactions between Pax6, Mafs, and retinoic acid nuclear receptors.31 This synergism could result from overlapping Pax6 and Maf-binding sites in the LSR1 region of the αB-promoter. Expression of αA-crystallin could be delayed due to the lack of cooperativity between Pax6 and Mafs as their binding sites are arranged differently from the mouse αB-crystallin gene (Figure 12). Levels of αA-crystallin expression in lens epithelium and lens fibers seem to be proportional to the levels of c-Maf expression.18,19 Expression of γF-crystallin is evident only later in primary lens fibers58 as Pax6 serves as an inhibitor of its expression mediated by a functional synergism between Maf and Sox proteins.31,59 It is also of interest that Pax6 has been established as a repressor of fiber-cell specific chicken βB1-crystallin competing with Maf proteins for the same region of DNA.15,60 In conclusion, different promoter architecture including mutual positions of Maf and Pax6-binding sites, their number, and their distances provide a number of mechanisms to mediate tissue-specific gene expression in the lens, pancreas, and likely in other tissues, during mammalian development.
Materials and Methods
Materials
The following oligonucleotides were synthesized by Invitrogen (Gaithersburg, MD) and were used as probes for EMSA: −113/ −91(5′-AGCTGCTGACGGTGCAGCCTCTC) −92/ −69(5′-TCCCCCGAGCTGAGCATAGACATT); −15/ +20(5′-GAACGCTAGCTCACCACCGCAC TGCCCAGAGGCTCCTG); +18/ +46(5′ -CTCCTGTCTGACTCACTGCCAGCCTTCGG). The oligonucleotides used as specific competitors: T-MARE (5′ -GCATTTGCTGACT CAGCATTTGGT); T-MARE-mutant (5′-GCATTTcagGACTCctgATTTGGT). Mutated nucleotides are indicated in lower case, and the consensus sequence of T-MARE is underlined.
Antibodies used for Western blots: anti-β-actin (1:2500, Abcam, Cambridge, UK), anti-Flag M2 monoclonal (1:2500, Sigma, St. Louis, MO); secondary antibodies were anti-mouse HRP (1:2000, Amersham, Piscataway, NJ). Antibodies used for ChIPs: anti-Pax6, anti-c-Maf, anti-TBP, and anti-Eya1 were purchased from Santa Cruz Biotechnology (St. Cruz, CA); anti-CREB-1 from Active-Motif (Carlsbad, CA), anti-H3 K9 acetylation from Upstate (Lake Placid, NY) and normal rabbit IgG from Oncogene Research Products (San Diego, CA).
Primers were designed using Primer 3†. PCR products were limited to 60–130 bp. Primers used for ChIPs were: αA-crystallin promoter (5′-AGTCATGTCGGGAAGACCTG and 5′-TGTGTGCTGGATGTGGTTCT;5′-CCCGAGCTGAGCATAGACAT and 5′-AGTCAGACAGGAGCCTCTGG); +4 kb downstream region of αA-crystallin promoter (5′-AGCAAGAAGGCAAGAGCAAA and 5′-GTCTAAGCTCCACCCCACAG); “negative” control promoter of ALASE (mouse erythroid-specific 5-aminolevulinate synthase) (5′-ATCGAATGAGTTAGTTTCACTACC and 5′-CCTTAAGGTATCTCAGGCCTCTGC).
The following oligonucleotides were used as primers for qRT-PCR analysis: Mouse c-Maf (forward 5′-GTGGTGGTGATGGCTCTTTT and reverse 5′-GTTACGGGGGAA TTCAGGTT); rat c-Maf (forward 5′ -CACACACACACACAGCAAGC and reverse 5′-TACAGGGGAATTCAGGTTGG); mouse MafB (forward 5′-GCCTCTTAGACT TGGGCAGA and reverse 5′-CCTTCCAGCTTGGAGAAAAG); mouse MafA (forward 5′-GCACCCGACTTCTTTCTGTG and reverse 5′ -GCCTGCGCAAACTTGTCC); mouse NRL (forward 5′-CTCAGTCCCAGAATGGCTTT, and reverse 5′-GAAGGCTCCCGCTTTATTTC); mouse and rat Pax6 (forward 5′ -GCACATGCAAACACACATGA and reverse 5′-ACTTGGACGGGAACTGACAC); mouse and rat Prox1 (5′-GCCCTCAACATGCACTACAA and reverse 5′-GGCATTGAAAAACTCCCGTA); mouse and rat CCNI (forward 5′-CATTCCTGATTGGCTTC CTC and reverse 5′-GTGGATCAACTGGGAGCTGT); mouse αA-crystallin (5′-GAGATTCACG GCAAACACAA and 5′-ACATTGGAAGGCAGACGGTA). Specificity of each primer set was determined using authentic cDNAs encoding both mouse and rat Mafs.
Database search to predict transcription factor binding sites
Transcription factor sites were predicted using on-line tools from The Transcription Factor DataBase (TRANSFAC)‡.
Plasmids, transfections and reporter assays
Mouse αA-crystallin gene promoter sequences −111 to +46, and truncated fragments −88/ +46, −111/ +26 and −88/ +26 were inserted into pGL-2-basic (Promega, Madison, WI). Single, double and triple mutations within the αA-crystallin −111/ +46 promoter were generated using the QuickChange site-directed mutagenesis kit (Stratagene, LaJolla, CA). Expression plasmids encoding Pax6, Pax6(5a), MafA, MafB, c-Maf and NRL were all driven by a CMV promoter as described earlier.30 Expression vectors containing 3x flag tagged Pax6, Pax6(5a), MafA, MafB, c-Maf and NRL were generated in p3xFLAG-CMV-10 (Sigma, St. Louis, MO). 5 μg of plasmid containing the αA-crystallin promoter −111/ +46 in pGL2 was co-transfected with Pax6 and Pax6(5a), MafA, MafB, c-Maf and NRL in different amounts into 293T cells as described in the Figure legends.30 To assess the activity of the −111 to +46 promoter region and the effect of deletions and specific mutations, 1 μg of αA-crystallin wild-type promoter and its specific mutants were transfected into αTN4-1 cells using Lipofectamine Plus Kit (Invitrogen). An internal control plasmid, pCMV Renilla luciferase, was included in all transfections. Firefly luciferase activities were normalized relative to the Renilla luciferase activity. Each transfection was conducted in triplicate at least twice.
Western blot analysis
The whole cell lysates of αTN4-1, N/N1003A and 293T cells containing 50–80 μg of total protein were loaded onto 12% (w/v) SDS-PAGE precast gels (Bio-Rad, Richmond, CA) and transferred to nitrocellulose membranes (Bio-Rad) as described.31 The membranes were incubated with the primary antibodies overnight at 4 °C.
Nuclear extracts, EMSA and DNase I footprinting
Four MARE-oligonucleotides −113/ −91, −92/ −69, −15/ +20 and +18/ +46 were radioactively end-labeled and incubated with the lens nuclear extracts prepared from the mouse lens epithelial cell line αTN4-1 as described earlier. Three oligonucleotides with Pax6-binding sites were −59/ −29 (site A), −88/ −56 (site B) and +18/ +46 (site C). EMSAs and antibody/EMSAs with rabbit antiPax6 polyclonal antibodies61 were performed as described.26,62 Specific oligonucleotide competitors (50 ng) were used at approximately 50-fold excess over the labeled probe. DNase I footprintings were performed as described.27
Chromatin immunoprecipitations (ChIPs)
70% confluent αTN4-1 cells were harvested and fixed by adding fresh formaldehyde to a final concentration of 1% at room temperature for ten minutes. Crosslinking was quenched by the addition of 2.5 M glycine solution to a final concentration of 0.125 M. Cells were lysed and sonicated to prepare chromatin of an average length of 500–600 bp. Antibody (2 μg) or rabbit IgG was incubated with blocked protein A and G beads (Sigma) overnight at 4 °C. Precleared chromatin (100 μl) was incubated with antibody bound protein A and G beads for five hours at 4 °C. Beads were washed three times and treated with Proteinase K at 55 °C for two to three hours followed by 65 °C overnight incubation to reverse crosslinking. Genomic DNA was finally eluted into 250 μl of water using QIAquick Spin Gel Purification Kit (Qiagen, Valencia, CA). Quantitative PCR reactions were performed as described below.38–40 The standard curve for each primer set was generated using 0.05, 0.2, 0.5 and 1% input DNA samples. The enrichments, given in relative input units, were obtained from Ct values and standard curves expressed relative to the 1% input. Enrichments using the control IgG were calculated separately. The experiments were performed using two independent chromatin preparations and each PCR reaction was performed in triplicate.
Quantitative RT-PCR
Total RNA was extracted from 70% confluent αTN4-1 mouse lens epithelial cells, one-day and one-week old CD-1 mouse lenses, micro-dissected two-day old rat lens epithelium and fiber cells using TRIzol (Invitrogen). RNA was treated with DNase I (Roche, Indianapolis, IN) at room temperature for 30 minutes followed by phenolchloroform extraction to avoid genomic DNA contamination. P0 and P4 total RNA from retina was obtained from Dr M. Dorrell (The Scripps Research Institute, La Jolla, CA). P22 total RNA from retina was obtained from Dr K. Mitton (Oakland Eye Institute, Rochester, MI). P14 total RNA from corneal epithelium was obtained from Dr T. Kays (National Eye Institute, Bethesda, MD). cDNAs were synthesized using SuperScriptII™ RNase H-RT (Invitrogen) according to the manufacturer's instructions and were diluted into a final volume of 250 μl with water. Diluted cDNA (2 μl) was mixed with 2 μM primers and 2x SYBG Mix (ABI, Foster City, CA) in a total volume of 8 μl. Samples were analyzed in a ABI 7900HT Sequence Detection System using a three-step protocol consisting of denaturation at 95 °C for 30 seconds, annealing at 60 °C for 30 seconds and extension at 72 °C for 30 seconds for 40 cycles. Data were analyzed using SDS2.0 (ABI) and Microsoft Excel (Redmond, WA). Series dilutions were used to determine primer efficiencies. Relative fold changes were then calculated using CCNI as an internal control as described.63
Supplementary Material
Supplementary Data
Supplementary data associated with this article can be found, in the online version, at doi:10.1016/j.jmb.2005.05.072
Acknowledgments
The authors thank Dr T. Stopka for advice on qChIPs and help with quantitative analysis, and Drs S. Saule and L. Wolf for critical reading of this manuscript. We are grateful to Drs M. Dorrell, T. Kays and K. Mitton for retinal and corneal RNA samples. We thank Drs M. Busslinger, J. A. Epstein, A. Sharma, A. Swaroop and K. Yoshida for expression clones or cDNAs used in this study. We thank Dr S. Saule for anti-Pax6 antibody and A. Cvekl, Jr. for his assistance in editing the manuscript. We also acknowledge AECOM DNA sequencing core facility and use of the ABI 7900HT equipment. The work was supported from NIH grants, EY12200 and 14237.
Abbreviations used
- qChIP
quantitative chromatin immunoprecipitation
- MARE
Maf responsive element
- qRT-PCR
quantitative reverse transcriptase-polymerase chain reaction
- EMSA
electrophoretic mobility shift assay
- PD
paired domain
- HD
homeodomain
Footnotes
References
- 1.Simpson TI, Price DJ. Pax6; a pleiotropic player in development. Bioessays. 2002;24:1041–1051. doi: 10.1002/bies.10174. [DOI] [PubMed] [Google Scholar]
- 2.van Heyningen V, Williamson KA. PAX6 in sensory development. Hum Mol Genet. 2002;11:1161–1167. doi: 10.1093/hmg/11.10.1161. [DOI] [PubMed] [Google Scholar]
- 3.Cvekl A, Tamm ER. Anterior eye development and ocular mesenchyme: new insights from mouse models and human diseases. Bioessays. 2004;26:374–386. doi: 10.1002/bies.20009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Donner AL, Maas RL. Conservation and non-conservation of genetic pathways in eye specification. Int J Dev Biol. 2004;48:743–753. doi: 10.1387/ijdb.041877ad. [DOI] [PubMed] [Google Scholar]
- 5.Gould DB, Smith RS, John SWM. Anterior segment development relevant to glaucoma. Int J Dev Biol. 2004;48:1015–1029. doi: 10.1387/ijdb.041865dg. [DOI] [PubMed] [Google Scholar]
- 6.Cvekl A, Yang Y, Chauhan BK, Cveklova K. Regulation of gene expression by Pax6 in ocular cells: a case of tissue-preferred expression of crystallins in lens. Int J Dev Biol. 2004;48:829–844. doi: 10.1387/ijdb.041866ac. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kondoh H, Uchikawa M, Kamachi Y. Interplay of Pax6 and SOX2 in lens development as a paradigm of genetic switch mechanisms for cell differentiation. Int J Dev Biol. 2004;48:819–827. doi: 10.1387/ijdb.041868hk. [DOI] [PubMed] [Google Scholar]
- 8.Reza HM, Yasuda K. Lens differentiation and crystallin regulation: a chick model. Int J Dev Biol. 2004;48:805–817. doi: 10.1387/ijdb.041863hr. [DOI] [PubMed] [Google Scholar]
- 9.Graw J. The genetic and molecular basis of congenital eye defects. Nature Rev Genet. 2003;4:876–888. doi: 10.1038/nrg1202. [DOI] [PubMed] [Google Scholar]
- 10.Bhat SP. Crystallins, genes and cataract. Prog Drug Res. 2003;60:205–262. doi: 10.1007/978-3-0348-8012-1_7. [DOI] [PubMed] [Google Scholar]
- 11.Robinson ML, Overbeek PA. Differential expression of alpha A- and alpha B-crystallin during murine ocular development. Invest Ophthalmol Vis Sci. 1996;37:2276–2284. [PubMed] [Google Scholar]
- 12.Haynes JI, 2nd, Duncan MK, Piatigorsky J. Spatial and temporal activity of the alpha B-crystallin/small heat shock protein gene promoter in transgenic mice. Dev Dyn. 1996;207:75–88. doi: 10.1002/(SICI)1097-0177(199609)207:1<75::AID-AJA8>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- 13.Wigle JT, Chowdhury K, Gruss P, Oliver G. Prox1 function is crucial for mouse lens-fibre elongation. Nature Genet. 1999;21:318–322. doi: 10.1038/6844. [DOI] [PubMed] [Google Scholar]
- 14.Duncan MK, Xie L, David LL, Robinson ML, Taube JR, Cui W, Reneker LW. Ectopic Pax6 expression disturbs lens fiber cell differentiation. Invest Ophthalmol Vis Sci. 2004;45:3589–3598. doi: 10.1167/iovs.04-0151. [DOI] [PubMed] [Google Scholar]
- 15.Duncan MK, Haynes JI, 2nd, Cvekl A, Piatigorsky J. Dual roles for Pax-6: a transcriptional repressor of lens fiber cell-specific beta-crystallin genes. Mol Cell Biol. 1998;18:5579–5586. doi: 10.1128/mcb.18.9.5579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ueda Y, Duncan MK, David LL. Lens proteomics: the accumulation of crystallin modifications in the mouse lens with age. Invest Ophthalmol Vis Sci. 2002;43:205–215. [PubMed] [Google Scholar]
- 17.Wawrousek EF, Chepelinsky AB, McDermott JB, Piatigorsky J. Regulation of the murine alpha A-crystallin promoter in transgenic mice. Dev Biol. 1990;137:68–76. doi: 10.1016/0012-1606(90)90008-7. [DOI] [PubMed] [Google Scholar]
- 18.Ring BZ, Cordes SP, Overbeek PA, Barsh GS. Regulation of mouse lens fiber cell development and differentiation by the Maf gene. Development. 2000;127:307–317. doi: 10.1242/dev.127.2.307. [DOI] [PubMed] [Google Scholar]
- 19.Kawauchi S, Takahashi S, Nakajima O, Ogino H, Morita M, Nishizawa M, et al. Regulation of lens fiber cell differentiation by transcription factor c-Maf. J Biol Chem. 1999;274:19254–19260. doi: 10.1074/jbc.274.27.19254. [DOI] [PubMed] [Google Scholar]
- 20.Nishiguchi S, Wood H, Kondoh H, Lovell-Badge R, Episkopou V. Sox1 directly regulates the gamma-crystallin genes and is essential for lens development in mice. Genes Dev. 1998;12:776–781. doi: 10.1101/gad.12.6.776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ho IC, Hodge MR, Rooney JW, Glimcher LH. The proto-oncogene c-maf is responsible for tissue-specific expression of interleukin-4. Cell. 1996;85:973–983. doi: 10.1016/s0092-8674(00)81299-4. [DOI] [PubMed] [Google Scholar]
- 22.Hedge SP, Kumar A, Kurschner C, Shapiro LH. c-Maf interacts with c-Myb to regulate transcription of an early myeloid gene during differentiation. Mol Cell Biol. 1998;18:2729–2737. doi: 10.1128/mcb.18.5.2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lyon MF, Jamieson RV, Perveen R, Glenister PH, Griffiths R, Boyd Y, et al. A dominant mutation within the DNA-binding domain of the bZIP transcription factor Maf causes murine cataract and results in selective alteration in DNA binding. Hum Mol Genet. 2003;12:585–594. [PubMed] [Google Scholar]
- 24.MacLean HE, Kim JI, Glimcher MJ, Wang J, Kronenberg HM, Glimcher LH. Absence of transcription factor c-maf causes abnormal terminal differentiation of hypertrophic chondrocytes during endochondral bone development. Dev Biol. 2003;262:51–63. doi: 10.1016/s0012-1606(03)00324-5. [DOI] [PubMed] [Google Scholar]
- 25.Ashery-Padan R, Marquardt T, Zhou X, Gruss P. Pax6 activity in the lens primordium is required for lens formation and for correct placement of a single retina in the eye. Genes Dev. 2000;14:2701–2711. doi: 10.1101/gad.184000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cvekl A, Kashanchi F, Sax CM, Brady JN, Piatigorsky J. Transcriptional regulation of the mouse alpha A-crystallin gene: activation dependent on a cyclic AMP-responsive element (DE1/CRE) and a Pax-6-binding site. Mol Cell Biol. 1995;15:653–660. doi: 10.1128/mcb.15.2.653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kantorow M, Cvekl A, Sax CM, Piatigorsky J. Protein–DNA interactions of the mouse alpha A-crystallin control regions. Differences between expressing and non-expressing cells. J Mol Biol. 1993;230:425–435. doi: 10.1006/jmbi.1993.1160. [DOI] [PubMed] [Google Scholar]
- 28.Ilagan JG, Cvekl A, Kantorow M, Piatigorsky J, Sax CM. Regulation of alphaA-crystallin gene expression. Lens specificity achieved through the differential placement of similar transcriptional control elements in mouse and chicken. J Biol Chem. 1999;274:19973–19978. doi: 10.1074/jbc.274.28.19973. [DOI] [PubMed] [Google Scholar]
- 29.Sax CM, Ilagan JG, Piatigorsky J. Functional redundancy of the DE-1 and alpha A-CRYBP1 regulatory sites of the mouse alpha A-crystallin promoter. Nucl Acids Res. 1993;21:2633–2640. doi: 10.1093/nar/21.11.2633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chauhan BK, Yang Y, Cveklova K, Cvekl A. Functional interactions between alternatively spliced forms of Pax6 in crystallin gene regulation and in haploinsufficiency. Nucl Acids Res. 2004;32:1696–1709. doi: 10.1093/nar/gkh334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yang Y, Chauhan BK, Cveklova K, Cvekl A. Transcriptional regulation of mouse alphaB-and gammaF-crystallin genes in lens: opposite promoter-specific interactions between Pax6 and large Maf transcription factors. J Mol Biol. 2004;344:351–368. doi: 10.1016/j.jmb.2004.07.102. [DOI] [PubMed] [Google Scholar]
- 32.Semenza GL. Transcription Factors and Human Disease Oxford Monographs on Medical Genetics No 37. Oxford University Press; Oxford: 1998. [Google Scholar]
- 33.Blank V, Andrews NC. The Maf transcription factors: regulators of differentiation. Trends Biochem Sci. 1997;22:437–441. doi: 10.1016/s0968-0004(97)01105-5. [DOI] [PubMed] [Google Scholar]
- 34.Yang Y, Cvekl A. Large Maf transcription factors: cousins of AP-1 proteins and important regulators of cellular differentiation. Einstein J Biol Med. 2005 doi: 10.23861/ejbm20072347. in the press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yoshida T, Yasuda K. Characterization of the chicken L-Maf, MafB and c-Maf in crystallin gene regulation and lens differentiation. Genes Cells. 2002;7:693–706. doi: 10.1046/j.1365-2443.2002.00548.x. [DOI] [PubMed] [Google Scholar]
- 36.Epstein J, Cai J, Glaser T, Jepeal L, Maas R. Identification of a Pax paired domain recognition sequence and evidence for DNA-dependent conformational changes. J Biol Chem. 1994;269:8355–8361. [PubMed] [Google Scholar]
- 37.Lengler J, Krausz E, Tomarev S, Prescott A, Quinlan RA, Graw J. Antagonistic action of Six3 and Prox1 at the gamma-crystallin promoter. Nucl Acids Res. 2001;29:515–526. doi: 10.1093/nar/29.2.515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rekhtman N, Choe KS, Matushansky I, Murray S, Stopka T, Skoultchi AI. PU.1 and pRB interact and cooperate to repress GATA-1 and block erythroid differentiation. Mol Cell Biol. 2003;23:7460–7474. doi: 10.1128/MCB.23.21.7460-7474.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Irvine RA, Hsieh CL. Q-PCR in combination with ChIP assays to detect changes in chromatin acetylation. Methods Mol Biol. 2004;287:45–52. doi: 10.1385/1-59259-828-5:045. [DOI] [PubMed] [Google Scholar]
- 40.Zavadil J, Cermak L, Soto-Nieves N, Bottinger EP. Integration of TGF-beta/Smad and Jagged1/Notch signalling in epithelial-to-mesenchymal transition. EMBO J. 2004;23:1155–1165. doi: 10.1038/sj.emboj.7600069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liu Q, Ji X, Breitman ML, Hitchcock PF, Swaroop A. Expression of the bZIP transcription factor gene Nrl in the developing nervous system. Oncogene. 1996;12:207–211. [PubMed] [Google Scholar]
- 42.Sakai M, Imaki J, Yoshida K, Ogata A, Matsushima-Hibaya Y, Kuboki Y, Nishizawa M, Nishi S. Rat maf related genes: specific expression in chondrocytes, lens and spinal cord. Oncogene. 1997;14:745–750. doi: 10.1038/sj.onc.1200869. [DOI] [PubMed] [Google Scholar]
- 43.Lecoin L, Sii-Felice K, Pouponnot C, Eychene A, Felder-Schmittbuhl MP. Comparison of maf gene expression patterns during chick embryo development. Gene Expr Patterns. 2004;4:35–46. doi: 10.1016/s1567-133x(03)00152-2. [DOI] [PubMed] [Google Scholar]
- 44.Kamachi Y, Uchikawa M, Tanouchi A, Sekido R, Kondoh H. Pax6 and SOX2 form a co-DNAbinding partner complex that regulates initiation of lens development. Genes Dev. 2001;15:1272–1286. doi: 10.1101/gad.887101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sander M, Neubuser A, Kalamaras J, Ee HC, Martin GR, German MS. Genetic analysis reveals that PAX6 is required for normal transcription of pancreatic hormone genes and islet development. Genes Dev. 1997;11:1662–1673. doi: 10.1101/gad.11.13.1662. [DOI] [PubMed] [Google Scholar]
- 46.Planque N, Leconte L, Coquelle FM, Benkhelifa S, Martin P, Felder-Schmittbuhl MP, Saule S. Interaction of Maf transcription factors with Pax-6 results in synergistic activation of the glucagon promoter. J Biol Chem. 2001;276:35751–35760. doi: 10.1074/jbc.M104523200. [DOI] [PubMed] [Google Scholar]
- 47.Kataoka K, Shioda S, Ando K, Sakagami K, Handa H, Yasuda K. Differentially expressed Maf family transcription factors, c-Maf and MafA, activate glucagon and insulin gene expression in pancreatic islet alpha- and beta-cells. J Mol Endocrinol. 2004;32:9–20. doi: 10.1677/jme.0.0320009. [DOI] [PubMed] [Google Scholar]
- 48.Brink C. Promoter elements in endocrine pancreas development and hormone regulation. Cell Mol Life Sci. 2003;60:1033–1048. doi: 10.1007/s00018-003-2247-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sivak JM, West-Mays JA, Yee A, Williams T, Fini ME. Transcription factors Pax6 and AP-2alpha interact to coordinate corneal epithelial repair by controlling expression of matrix metalloproteinase gelatinase B. Mol Cell Biol. 2004;24:245–257. doi: 10.1128/MCB.24.1.245-257.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kozmik Z, Daube M, Frei E, Norman B, Kos L, Dishaw LJ, et al. Role of Pax genes in eye evolution: a cnidarian PaxB gene uniting Pax2 and Pax6 functions. Dev Cell. 2003;5:773–785. doi: 10.1016/s1534-5807(03)00325-3. [DOI] [PubMed] [Google Scholar]
- 51.Carosa E, Kozmik Z, Rall JE, Piatigorsky J. Structure and expression of the scallop Omega-crystallin gene. Evidence for convergent evolution of promoter sequences. J Biol Chem. 2002;277:656–664. doi: 10.1074/jbc.M107004200. [DOI] [PubMed] [Google Scholar]
- 52.Zhao H, Yang Y, Rizo CM, Overbeek PA, Robinson ML. Insertion of a Pax6 consensus binding site into the alphaA-crystallin promoter acts as a lens epithelial cell enhancer in transgenic mice. Invest Ophthalmol Vis Sci. 2004;45:1930–1939. doi: 10.1167/iovs.03-0856. [DOI] [PubMed] [Google Scholar]
- 53.Sakai M, Serria MS, Ikeda H, Yoshida K, Imaki J, Nishi S. Regulation of c-maf gene expression by Pax6 in cultured cells. Nucl Acids Res. 2001;29:1228–1237. doi: 10.1093/nar/29.5.1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ogino H, Yasuda K. Induction of lens differentiation by activation of a bZIP transcription factor, L-Maf. Science. 1998;280:115–118. doi: 10.1126/science.280.5360.115. [DOI] [PubMed] [Google Scholar]
- 55.Reza HM, Ogino H, Yasuda KLM. Maf, a downstream target of Pax6, is essential for chick lens development. Mech Dev. 2002;116:61–73. doi: 10.1016/s0925-4773(02)00137-5. [DOI] [PubMed] [Google Scholar]
- 56.Duncan MK, Cvekl A, Kantorow M, Piatigorsky J. Lens crystallins. In: Lovicu FJ, Robinson ML, editors. Development of the Ocular Lens. Cambridge University Press; 2004. pp. 119–150. [Google Scholar]
- 57.Reza HM, Yasuda K. Roles of Maf family proteins in lens development. Dev Dyn. 2004;229:440–448. doi: 10.1002/dvdy.10467. [DOI] [PubMed] [Google Scholar]
- 58.Goring DR, Rossant J, Clapoff S, Breitman ML, Tsui LC. In situ detection of betagalactosidase in lenses of transgenic mice with a gamma-crystallin/lacZ gene. Science. 1987;235:456–458. doi: 10.1126/science.3099390. [DOI] [PubMed] [Google Scholar]
- 59.Rajaram N, Kerppola TK. Synergistic transcription activation by Maf and Sox and their subnuclear localization are disrupted by a mutation in Maf that causes cataract. Mol Cell Biol. 2004;24:5694–5709. doi: 10.1128/MCB.24.13.5694-5709.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Cui W, Tomarev SI, Piatigorsky J, Chepelinsky AB, Duncan MK. Mafs, Prox1, and Pax6 can regulate chicken betaB1-crystallin gene expression. J Biol Chem. 2004;279:11088–11095. doi: 10.1074/jbc.M312414200. [DOI] [PubMed] [Google Scholar]
- 61.Carriere C, Plaza S, Martin P, Quatannens B, Bailly M, Stehelin D, Saule S. Characterization of quail Pax-6 (Pax-QNR) proteins expressed in the neuroretina. Mol Cell Biol. 1993;13:7257–7266. doi: 10.1128/mcb.13.12.7257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chauhan BK, Yang Y, Cveklova K, Cvekl A. Functional properties of natural human PAX6 and PAX6(5a) mutants. Invest Ophthalmol Vis Sci. 2004;45:385–392. doi: 10.1167/iovs.03-0968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 64.Chen Q, Dowhan DH, Liang D, Moore DD, Overbeek PA. CREB-binding protein/p300 co-activation of crystallin gene expression. J Biol Chem. 2002;277:24081–24089. doi: 10.1074/jbc.M201821200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Data
Supplementary data associated with this article can be found, in the online version, at doi:10.1016/j.jmb.2005.05.072